Abstract
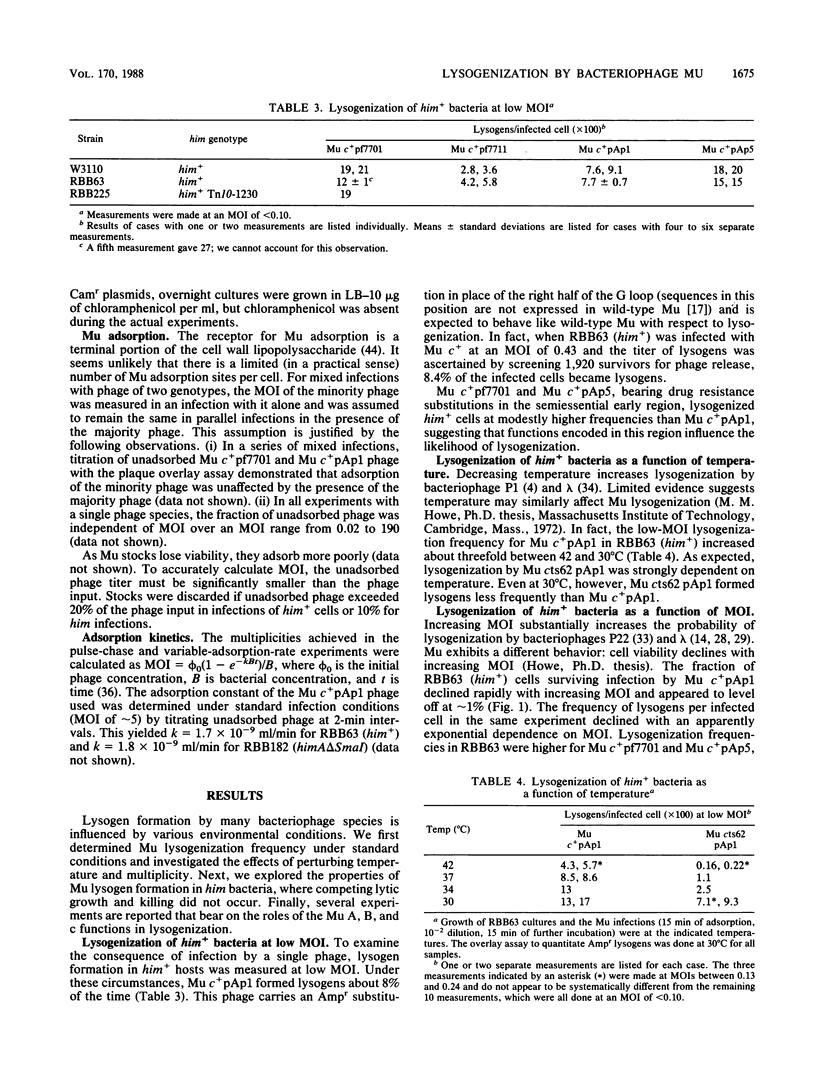
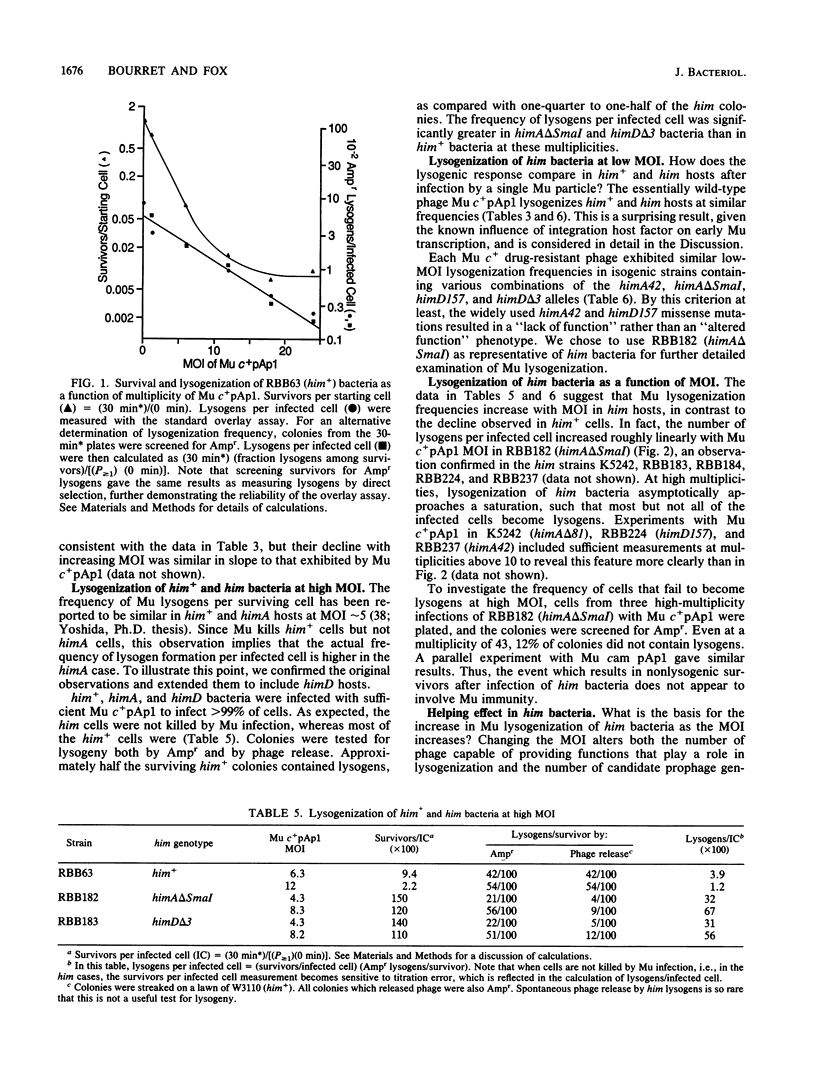
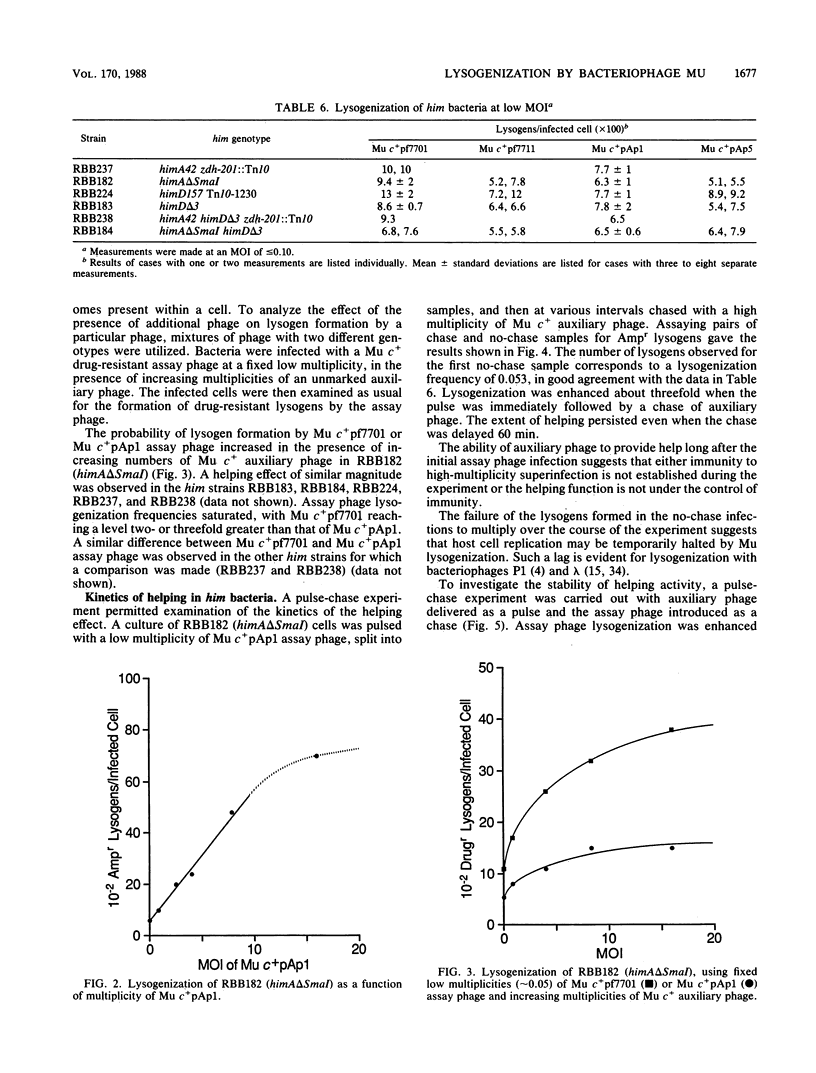
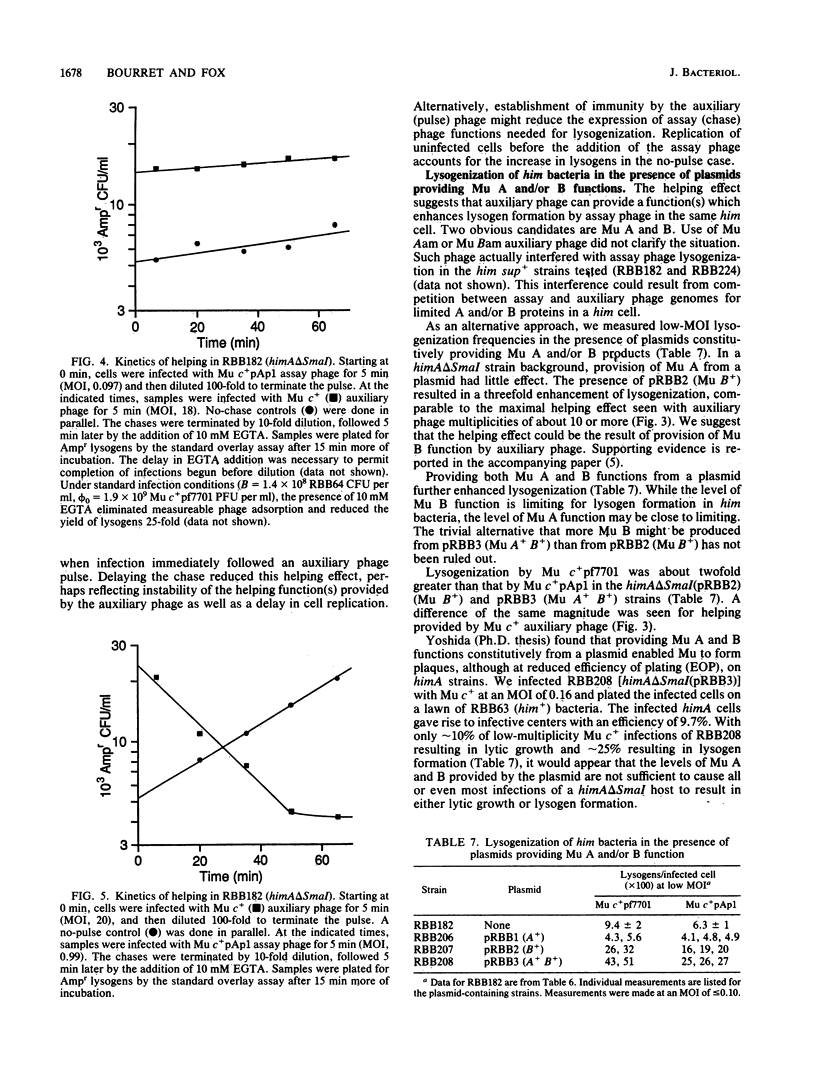
The possible outcomes of infection of Escherichia coli by bacteriophage Mu include lytic growth, lysogen formation, nonlysogenic surviving cells, and perhaps simple killing of the host. The influence of various parameters, including host himA and himD mutations, on lysogeny and cell survival is described. Mu does not grow lytically in or kill him bacteria but can lysogenize such hosts. Mu c+ lysogenizes about 8% of him+ bacteria infected at low multiplicity at 37 degrees C. The frequency of lysogens per infected him+ cell diminishes with increasing multiplicity of infection or with increasing temperature over the range from 30 to 42 degrees C. In him bacteria, the Mu lysogenization frequency increases from about 7% at low multiplicity of infection to approach a maximum where most but not all cells are lysogens at high multiplicity of infection. Lysogenization of him hosts by an assay phage marked with antibiotic resistance is enhanced by infection with unmarked auxiliary phage. This helping effect is possible for at least 1 h, suggesting that Mu infection results in formation of a stable intermediate. Mu immunity is not required for lysogenization of him hosts. We argue that in him bacteria, all Mu genomes which integrate into the host chromosome form lysogens.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akroyd J., Barton B., Lund P., Maynard Smith S., Sultana K., Symonds N. Mapping and properties of the gam and sot genes of phage mu: their possible roles in recombination. Cold Spring Harb Symp Quant Biol. 1984;49:261–266. doi: 10.1101/sqb.1984.049.01.030. [DOI] [PubMed] [Google Scholar]
- BERTANI G. Lysogeny. Adv Virus Res. 1958;5:151–193. doi: 10.1016/s0065-3527(08)60673-9. [DOI] [PubMed] [Google Scholar]
- BERTANI G., NICE S. J. Studies on lysogenesis. II. The effect of temperature on the lysogenization of Shigella dysenteriae with phage P1. J Bacteriol. 1954 Feb;67(2):202–209. doi: 10.1128/jb.67.2.202-209.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Fox M. S. Intermediates in bacteriophage Mu lysogenization of Escherichia coli him hosts. J Bacteriol. 1988 Apr;170(4):1683–1690. doi: 10.1128/jb.170.4.1683-1690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G., Giddens E. B., Miller J. L., Gloor G. A truncated form of the bacteriophage Mu B protein promotes conservative integration, but not replicative transposition, of Mu DNA. Cell. 1985 Jul;41(3):857–865. doi: 10.1016/s0092-8674(85)80066-0. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Gloor G., Miller J. L., Kennedy D. L., Giddens E. B., Nagainis C. R. Transposition of bacteriophage mu DNA: expression of the A and B proteins from lambda pL and analysis of infecting mu DNA. Cold Spring Harb Symp Quant Biol. 1984;49:279–284. doi: 10.1101/sqb.1984.049.01.033. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Kennedy D. L., Evans D. Predominant integration end products of infecting bacteriophage Mu DNA are simple insertions with no preference for integration of either Mu DNA strand. Virology. 1983 Jul 15;128(1):48–59. doi: 10.1016/0042-6822(83)90317-3. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Kahmann R., Kamp D. Electron microscopic characterization of DNAs of non-defective deletion mutants of bacteriophage Mu. J Mol Biol. 1977 Jul 15;113(4):591–609. doi: 10.1016/0022-2836(77)90224-8. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- FRY B. A. Conditions for the infection of Escherichia coli with lambda phage and for the establishment of lysogeny. J Gen Microbiol. 1959 Dec;21:676–684. doi: 10.1099/00221287-21-3-676. [DOI] [PubMed] [Google Scholar]
- FRY B. A., GROS F. The metabolic activities of Escherichia coli during the establishment of lysogeny. J Gen Microbiol. 1959 Dec;21:685–692. doi: 10.1099/00221287-21-3-685. [DOI] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Goosen T., van Meeteren A., Wijffelman C., van de Putte P. Properties of the recombinant plasmid pGP1 containing part of the early region of bacteriophage mu. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1179–1185. doi: 10.1101/sqb.1979.043.01.133. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Plasterk R. H., van de Putte P. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature. 1982 May 27;297(5864):339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- Goosen N., van Heuvel M., Moolenaar G. F., van de Putte P. Regulation of Mu transposition. II. The escherichia coli HimD protein positively controls two repressor promoters and the early promoter of bacteriophage Mu. Gene. 1984 Dec;32(3):419–426. doi: 10.1016/0378-1119(84)90017-9. [DOI] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Role of ner protein in bacteriophage Mu transposition. J Bacteriol. 1986 Aug;167(2):503–507. doi: 10.1128/jb.167.2.503-507.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Schumm J. W., Taylor A. L. The S and U genes of bacteriophage mu are located in the invertible G segment of mu DNA. Virology. 1979 Jan 15;92(1):108–124. doi: 10.1016/0042-6822(79)90218-6. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Knight D. M., Das A., Miller H. I., Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982 Dec;31(3 Pt 2):565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Structure of inserted bacteriophage Mu-1 DNA and physical mapping of bacterial genes by Mu-1 DNA insertion. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2823–2827. doi: 10.1073/pnas.69.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., Fox M. S. A genetic analysis of primary products of bacteriophage lambda recombination. Genetics. 1986 Mar;112(3):409–420. doi: 10.1093/genetics/112.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A., Flamm E., Weisberg R. A. An Escherichia coli mutant unable to support site-specific recombination of bacteriophage lambda. J Mol Biol. 1985 May 25;183(2):129–140. doi: 10.1016/0022-2836(85)90207-4. [DOI] [PubMed] [Google Scholar]
- Knoll B. J. Isolation and characterization of mutations in the cIII gene of bacteriophage lambda which increase the efficiency of lysogenization of Escherichia coli K-12. Virology. 1979 Jan 30;92(2):518–531. doi: 10.1016/0042-6822(79)90154-5. [DOI] [PubMed] [Google Scholar]
- Kourilsky P. Lysogenization by bacteriophage lambda. I. Multiple infection and the lysogenic response. Mol Gen Genet. 1973 Apr 12;122(2):183–195. doi: 10.1007/BF00435190. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Krause H. M., Rothwell M. R., Higgins N. P. The early promoter of bacteriophage Mu: definition of the site of transcript initiation. Nucleic Acids Res. 1983 Aug 25;11(16):5483–5495. doi: 10.1093/nar/11.16.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVINE M. Mutations in the temperate phage P22 and lysogeny in Salmonella. Virology. 1957 Feb;3(1):22–41. doi: 10.1016/0042-6822(57)90021-1. [DOI] [PubMed] [Google Scholar]
- LIEB M. The establishment of lysogenicity in Escherichia coli. J Bacteriol. 1953 Jun;65(6):642–651. doi: 10.1128/jb.65.6.642-651.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D., Symonds N. The isolation and characterisation of a plaque-forming derivative of bacteriophage Mu carrying a fragment of Tn3 conferring ampicillin resistance. Mol Gen Genet. 1979 May 4;172(2):179–184. doi: 10.1007/BF00268280. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Khatoon H., DuBow M., Ambrosio L., De Bruijn F., Bukhari A. I. Integration of bacteriophage mu DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1151–1158. doi: 10.1101/sqb.1979.043.01.130. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Friedman D. I. An E. coli gene product required for lambda site-specific recombination. Cell. 1980 Jul;20(3):711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kikuchi A., Nash H. A., Weisberg R. A., Friedman D. I. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Miller H. I. Multilevel regulation of bacteriophage lambda lysogeny by the E. coli himA gene. Cell. 1981 Jul;25(1):269–276. doi: 10.1016/0092-8674(81)90252-x. [DOI] [PubMed] [Google Scholar]
- O'Day K., Schultz D., Ericsen W., Rawluk L., Howe M. Correction and refinement of the genetic map of bacteriophage Mu. Virology. 1979 Mar;93(2):320–328. doi: 10.1016/0042-6822(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Patterson T. A., Martin K. A., Bukhari A. I. Analysis of the regulation of the transposition functions of bacteriophage mu by using gene fusions. Cold Spring Harb Symp Quant Biol. 1984;49:267–272. doi: 10.1101/sqb.1984.049.01.031. [DOI] [PubMed] [Google Scholar]
- Ross W., Shore S. H., Howe M. M. Mutants of Escherichia coli defective for replicative transposition of bacteriophage Mu. J Bacteriol. 1986 Sep;167(3):905–919. doi: 10.1128/jb.167.3.905-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandulache R., Prehm P., Kamp D. Cell wall receptor for bacteriophage Mu G(+). J Bacteriol. 1984 Oct;160(1):299–303. doi: 10.1128/jb.160.1.299-303.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaus N. A., Wright A. Inhibition of Escherichia coli exonuclease V by bacteriophage Mu. Virology. 1980 Apr 15;102(1):214–217. doi: 10.1016/0042-6822(80)90083-5. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L. BACTERIOPHAGE-INDUCED MUTATION IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1963 Dec;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti F., Barksdale C., Abelson J. Mu-1 bacteriophage DNA. Virology. 1970 Jul;41(3):567–568. doi: 10.1016/0042-6822(70)90179-0. [DOI] [PubMed] [Google Scholar]
- Van Leerdam E., Karreman C., van de Putte P. Ner, a cro-like function of bacteriophage Mu. Virology. 1982 Nov;123(1):19–28. doi: 10.1016/0042-6822(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Waggoner B. T., Marrs C. F., Howe M. M., Pato M. L. Multiple factors and processes involved in host cell killing by bacteriophage Mu: characterization and mapping. Virology. 1984 Jul 15;136(1):168–185. doi: 10.1016/0042-6822(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Weil J., Cunningham R., Martin R., 3rd, Mitchell E., Bolling B. Characteristics of lambda p4, a lambda derivative containing 9 per cent excess DNA. Virology. 1972 Nov;50(2):373–380. doi: 10.1016/0042-6822(72)90388-1. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- van Meeteren R., Giphart-Gassler M., van de Putte P. Transcription of bacteriophage Mu. II. Transcription of the repressor gene. Mol Gen Genet. 1980;179(1):185–189. doi: 10.1007/BF00268462. [DOI] [PubMed] [Google Scholar]



