Abstract
Telomerase is a eukaryotic reverse transcriptase that adds simple sequence repeats to chromosome ends by copying a template sequence within the RNA component of the enzyme. We describe here the identification of a Tetrahymena telomerase protein with reverse transcriptase motifs, p133. This subunit is associated with the previously identified Tetrahymena telomerase RNA and the telomerase proteins p80 and p95 in immunoprecipitation assays. Therefore, all four known Tetrahymena telomerase components are present in a single complex. Expressed in rabbit reticulocyte lysate, recombinant p133 and telomerase RNA alone catalyze a reverse transcriptase activity with some similarities to and some differences from native Tetrahymena telomerase. These experiments suggest a complexity of telomerase structure and function.
Telomeres in most organisms are comprised of tandem simple sequence repeats (see ref. 1 for review). The total length of telomeric repeat sequence at each chromosome end is determined in a balance of sequence loss and sequence addition. One major influence on telomere length is the enzyme telomerase. The telomerase ribonucleoprotein complex (RNP) can extend a DNA 3′ end by reverse transcribing a template region within its RNA component (2, 3). Telomerase from at least some species also can catalyze the template-directed nucleolytic cleavage of primer or product polynucleotides (4–6). In vitro, telomerase has a general preference for single-stranded, G-rich oligonucleotide primers (see ref. 7 for review). Although primer elongation is enhanced by hybridization of a primer 3′ end with the template, primer affinity derives in large part from interaction with protein-based anchor site(s). Product interaction with the anchor site is required for the highly processive elongation catalyzed by Tetrahymena telomerase, in which repeats from multiple rounds of template copying are added to an individual substrate before dissociation (8).
The molecular basis for the biochemical properties of telomerase is not fully understood. The RNA component of telomerase has been identified in numerous species (see ref. 1 for review). Telomerase protein components have been identified in ciliates (Tetrahymena thermophila p80, p95; Euplotes aediculatus p123), yeast (S. cerevisiae Est2p), and mammalian cells [TP1/TLP1, telomerase reverse transcriptase (TERT)] (see ref. 9 for review). The evolutionary conservation of telomerase RNA and protein sequences is strikingly low. The only obvious sequence motif other than the RNA template is a reverse transcriptase active site in the TERT protein (10). Also, only limited biochemical characterization of telomerase proteins has been described. Two ciliate proteins have been studied in purified, recombinant form: Tetrahymena p80 and p95 interact with each other and with telomerase RNA, and the p95 subunit interacts with single-stranded telomeric DNA in a sequence-specific, length-dependent manner (11). TERT genes have been studied by expressing mutant forms in vivo: site-specific substitutions in polymerase motif regions produce cells or extracts defective in telomerase activity (10). In addition, constitutive expression of human TERT is sufficient to activate telomerase in a previously telomerase-negative cell, and expression of human TERT and telomerase RNA by coupled in vitro transcription/translation in reticulocyte lysate is sufficient to produce reverse transcriptase activity (12–15). Finally, coimmunoprecipitation studies from human cells indicate that TP1 and TERT subunits interact (16).
In this report, we describe the cloning of a Tetrahymena telomerase protein subunit from the TERT family, which we refer to as p133. We show that Tetrahymena p133, p95, p80, and telomerase RNA exist as a stable complex. In addition, we characterize the activities of recombinant p133 produced in rabbit reticulocyte lysate. Lysate expressing p133 and telomerase RNA can catalyze reverse transcription and nucleolytic cleavage, but the observed activities only partially reconstitute native Tetrahymena telomerase. Our findings suggest that telomerase functionrequires a coordination of separable biochemical activities in the RNP.
MATERIALS AND METHODS
Cloning of the p133 Gene.
Mated T. thermophila total RNA was reverse transcribed with an anchor-T primer (GCTGAATTCGTCGACATCGATTTTTTTTTTTTTTTTT). This cDNA was used in PCR with TC primer [TCGAATTCTT(TC)TT(TC)TA(TC)(GT)(GT)(ATC)AC(ATC)GA] according to the following protocol: 4 cycles (94°C, 45 s; 35°C, 45 s; to 72°C 1°C/s; 72°C 2 min); add anchor-T primer; continue 35 cycles (94°C, 30 s; 50°C, 30 s; 72°C 2 min). Product DNA was amplified in a second round nested PCR under the same conditions with TG primer [(TC)A(GA)AAGCTTGG(TAC)AT(ATC)CC(ATC)(CT)A(AG)GG] and TB2 primer [GTGAATTC(AT)A(AG)(ACGT)A(AG)(AG)(TA)A(AG)TC(AG)TC]. One additional degenerate PCR used TC primer with a nested primer set derived from PCR product sequence. Additional T. thermophila cDNA sequence was obtained by RT-PCR and rapid amplification of cDNA ends (RACE). To isolate T. thermophila genomic sequence, macronuclear DNA was amplified with gene-specific primers and a genomic DNA library was amplified by using one gene-specific primer and one vector primer. Sequence alignments were done with Genetics Computer Group (GCG) bestfit.
Immunoprecipitation.
Anti-Ct p133 antibody was raised against the C-terminal peptide CPKISAKSNQQNTN. Anti-AB p133 antibody was raised against the peptide CDDQILQKGFKEIQSDDRP between sequence motifs A and B. Polyclonal rabbit serum was raised against each peptide conjugated to keyhole limpet hemacyanin and was purified on peptide coupled to thiol Sepharose. Antibody against p80 used for immunoprecipitation was raised and affinity-purified against recombinant p80 expressed in Escherichia coli and will be described elsewhere. The control antibody used in Figs. 2 and 3 was an anti-peptide antibody raised and affinity-purified in the same manner as the p133 antibodies; other polyclonal antibodies were used with the same results. Purified antibody was pre-bound to Protein A–Sepharose at ≈1 μg/μl and was washed in PBS followed by IP wash buffer (20 mM Tris⋅HCl, pH 8.0/0.1 M NaCl/1 mM MgCl2/10% glycerol/0.1% Nonidet P-40/0.1 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin).
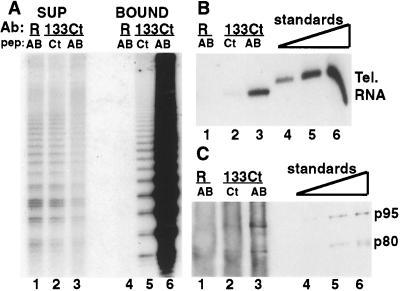
Figure 2.
Immunoprecipitation with p133 antibody from whole cell extract. (A) Telomerase activity was assayed in supernatants (lanes 1–3) or immunoprecipitates resuspended at 30-fold greater relative concentration (lanes 4–6). Protein A-bound anti-Ct p133 antibody was incubated with extract in the presence of Ct competitor peptide (lanes 2 and 5) or nonspecific peptide AB (lanes 3 and 6), or Protein A-bound random control antibody (R) was incubated with extract in the presence of nonspecific peptide AB (lanes 1 and 4). (B) Telomerase RNA in immunoprecipitates was analyzed by Northern blot. The same volume of bound fractions as in (A) was analyzed (lanes 1–3) in comparison with recombinant telomerase RNA standards (lanes 4–6: 0.1, 0.3, 1.0 ng RNA). (C) Telomerase proteins in immunoprecipitates were analyzed by immunoblot. Three times the volume of bound fraction in (A) was probed for p80 and p95 (lanes 1–3) in comparison with recombinant protein standards (lanes 4–6: 0.25, 1.0, 4.0 ng each protein). The high background in the bound fractions derives from recognition of the primary antibody used for immunoprecipitation.
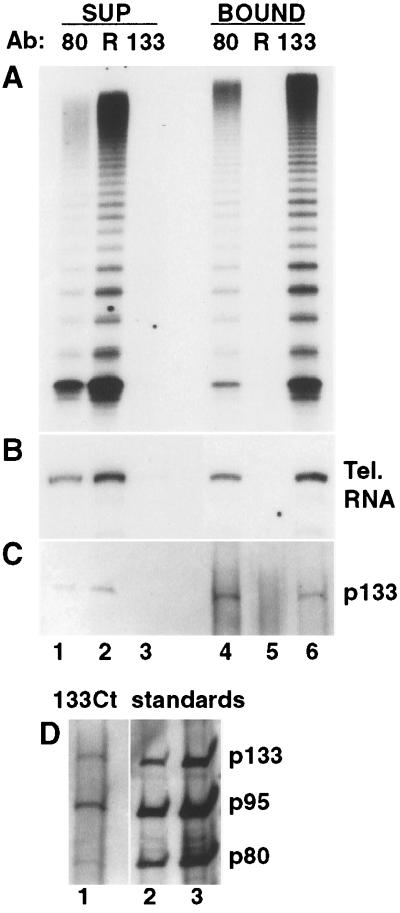
Figure 3.
Immunoprecipitation of p133 from partially purified telomerase fractions. (A–C) Equal relative volumes of supernatants (lanes 1–3) or immunoprecipitates (lanes 4–6) were assayed for telomerase activity (A) or for telomerase RNA by Northern blot (B). Supernatants and an ≈5-fold greater relative amount of immunoprecipitates were analyzed for p133 by immunoblot (C). Protein A-bound polyclonal anti-p80 antibody (lanes 1 and 4), random control antibody (R; lanes 2 and 5), or anti-Ct p133 antibody (lanes 3 and 6) was incubated with extract. The supernatants and immunoprecipitates shown in B were run on separate gels. (D) The immunoprecipitate from a complete activity depletion with anti-Ct p133 antibody was examined by immunoblot for p133, p95, and p80 (lane 1) in comparison with recombinant protein standards (lanes 2–3: 0.25, 1.0 ng each protein, not normalized for relative mass).
Mated cells were used for whole cell extract immunoprecipitation because they have maximal telomerase activity and less background radiolabeling activity. Strains CU428 and B2086 were grown to mid-log phase, starved, and then mated for ≈8 h before harvesting. Cells were lysed in T2MG buffer (20 mM Tris⋅HCl, pH 8.0/1 mM MgCl2/10% glycerol) with 0.2% Nonidet P-40 and centrifuged at 40,000 rpm for 50 min in a Ti45 rotor. Starved cell extracts were the source for partially purified telomerase, using previously described protocols to concentrate and enrich for the RNP (17). Immunoprecipitation samples were adjusted to 0.1 M NaCl and 0.1–0.2% Nonidet P-40 in T2MG, with 25 μg/ml peptide if indicated, before addition of antibody-bound Protein A–Sepharose (1:10–1:60 ratio of bead slurry to extract, ≈50-fold molar excess of peptide over antibody). Binding reactions were mixed end-over-end at 4°C, washed 4 times each in 50 volumes of IP wash buffer, and then resuspended in IP wash buffer to 1/10–1/30th the initial volume of sample.
Telomerase Activity, Proteins, and RNA.
A recombinant p133 coding region was designed based on optimized codon usage (sequence available on request). Approximately 90-nt oligonucleotides with approximately 16-bp terminal overlaps were annealed pairwise and extended to create double-stranded DNA. Pools of extended pairs, corresponding to segments between unique restriction sites, were subject to PCR with flanking, shorter, segment-end primers. Product DNA was purified, digested with appropriate restriction enzymes, cloned, and sequenced. Individual segments then were combined to create the full length coding region. The double aspartic acid mutant (in motif C, DD changed to AA) was made by using site-directed mutagenesis as described (11). Proteins used for concentration standards were expressed in E. coli from pRSET (Invitrogen).
Recombinant protein for activity assays was expressed from pCITE-4a (Invitrogen) by using a T7 coupled transcription/translation system (Promega) with [35S]methionine. Telomerase RNA was transcribed from Fok I-digested template DNA (18). Where indicated, product RNA was purified from T7 RNA polymerase transcription reactions (Ambion) after DNase treatment by organic extraction and precipitation. Template RNAs T1, T2, and T3 were made by chemical synthesis according to standard methods (Applied Biosystems International). For activity assays, samples were diluted into 10 μl of T2MG and combined with 10 μl of reaction mix. Unless indicated otherwise, all reactions contained 0.5 μM primer, 500 μM dTTP, and 0.3 μM 32P-dGTP, were performed at 30°C for approximately 1 h; and were analyzed on 9–10% (19:1) acrylamide, 7 M urea, 0.6X TBE gels. Immunoprecipitation reactions were assayed with primer d(T2G4)3. Activity assays comparing recombinant and purified enzymes were supplemented with an additional 2.5 mM MgCl2 and 500 μM of any other nucleotides as indicated.
For immunoblots, proteins were resolved by SDS/PAGE and then transferred to nitrocellulose in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) (pH 11.0)/10% methanol at 0.5 A for 1.1 h. Affinity-purified rabbit polyclonal primary antibodies were used. The p133 was detected by anti-Ct or anti-AB, p95 was detected by 988 (raised against denatured recombinant N-terminal domain of p95), and p80 was detected by 1589 (raised against denatured recombinant C-terminal domain of p80). Secondary antibody was a horseradish peroxidase-conjugated anti-rabbit (Bio-Rad) detected by enhanced chemiluminescence (Amersham). For Northern analysis, RNA was purified by organic extraction and precipitation. RNAs were resolved on 6% (19:1) acrylamide, 7 M urea, 0.6X TBE gels, transferred to Hybond-N+ (Amersham), and hybridized in 4X Denhardt’s solution, 6X SSC, and 0.1% SDS. Telomerase RNA was detected with a 32P-end-labeled oligonucleotide complementary to the template region or template-adjacent sequence.
RESULTS
Identification of Tetrahymena thermophila TERT, p133.
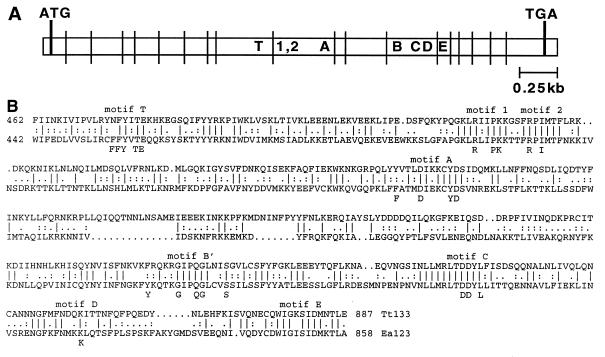
To identify Tetrahymena thermophila cDNAs that contain sequence motifs common to the TERT family (19), a degenerate PCR strategy was adopted. After numerous protocols, a product DNA was obtained with more than a minimal sequence homology to E. aediculatus p123 (10). Using RT-PCR, 5′ and 3′ RACE, and PCR from genomic libraries and genomic DNA, the cDNA coding region and corresponding genomic region were identified and sequenced (Fig. 1). The 3,351-bp ORF is predicted to encode a protein product of 133 kDa. It contains 68 TAA and TAG codons that specify glutamine in Tetrahymena. This ORF is likely to be complete. The indicated start codon ATG follows an in-frame stop codon in the 5′ UTR (a second ATG codon closely follows the first); expression of recombinant p133 results in protein with the same electrophoretic mobility as a crossreactive Tetrahymena polypeptide (see below); and 5′ and 3′ UTRs typical in length and sequence were identified (data not shown). The coding region is interrupted by 18 introns (Fig. 1A), a large number for a Tetrahymena macronuclear gene. The introns are of typical size for Tetrahymena (47–296 bp), are A/T rich, and contain consensus splice site sequences (20). The overall level of homology between ciliate TERT proteins is not high (29% identity, 53% similarity). However, relative to nonciliate TERT family members, T. thermophila and E. aediculatus proteins demonstrate a much greater sequence homology in the reverse transcriptase motif region (Fig. 1B; 38% identity, 61% similarity). Most or all of these previously identified sequence motifs (10, 19) are likely to form the polymerase active site. In particular, the aspartic acids of motifs A and C are expected to play a major role by binding the metal ions that participate in catalysis.
Figure 1.
The cDNA and genomic region encoding p133. (A) The cDNA sequence of p133 is indicated as an open box. Locations of the predicted start codon (ATG) and stop codon (TGA) are shown by thick lines. Locations of the 18 introns removed from the cDNA are shown by thin lines. Conserved sequence motifs are lettered and numbered. (B) The motif region sequence of Tetrahymena thermophila p133 (Tt133, Top) and Euplotes aediculatus p123 (Ea123, Bottom; ref. 10) are aligned over the indicated amino acids. Motif designations are given above the alignment, and the previously derived consensus (19) is given below.
Interaction of p133, Telomerase RNA, and p80/p95.
To investigate the association of p133 with telomerase activity, we raised antibodies against peptides from within the predicted coding region. Affinity-purified p133 antibody (anti-Ct) bound to Protein A–Sepharose could deplete telomerase activity from a whole cell extract with a corresponding recovery of activity in the bound fraction (Fig. 2A). Immunoprecipitation was efficient in the presence of a nonspecific competitor peptide (AB peptide; Fig. 2A, lanes 3 and 6) but was reduced by the presence of specific competitor peptide in the binding reaction (Ct peptide; Fig. 2A, lanes 2 and 5) and was not observed with random control antibodies raised and affinity-purified in a similar manner (R antibody; Fig. 2A, lanes 1 and 3). We next investigated the association of p133 with p80, p95, and telomerase RNA to determine whether all four components formed parts of a single RNP. Telomerase RNA and p80 and p95 were recovered in the anti-Ct bound fraction, competed by specific peptide (Fig. 2 B and C, lane 2) but not nonspecific peptide (Fig. 2 B and C, lane 3), and not observed in a control antibody immunoprecipitation (Fig. 2 B and C, lane 1). Using recombinant standards of known RNA or protein concentration (Fig. 2 B and C, lanes 4–6) and correcting for relative sample volumes and sample treatments, approximately equivalent ratios of RNA, p80, and p95 were recovered after p133 immunoprecipitation. A second p133 antibody (anti-AB) also immunoprecipitated p80, p95, and telomerase RNA; this antibody depleted activity from the extract, but no activity was recovered in the immunoprecipitate (data not shown).
We also examined the interaction of p133, p80, and p95 in a partially purified telomerase fraction representative of the preparations used for enzyme activity characterization (17). We assayed the coimmunoprecipitation of p133 with p80/p95 by using an antibody against p80. Although the interaction of p80 antibody with telomerase RNP is not as efficient as its interaction with p80 alone, because of the polyclonal nature of the antibody, it has the advantage of immunoprecipitating telomerase in active form. Equal mass amounts of p80 and p133 anti-Ct peptide antibodies bound to Protein A–Sepharose depleted telomerase activity from a partially purified fraction whereas a random control antibody did not (Fig. 3A, lanes 1–3). Activity was recovered in both p80 and p133 immunoprecipitates but not in the control (Fig. 3A, lanes 4–6). Both p80 and p133 antibodies were capable of quantitatively depleting telomerase activity if used in sufficient amount, although the p133 anti-Ct antibody was more efficient (Fig. 3A).
As demonstrated previously (17) with a different p80 antibody and above in whole cell extract with p133 antibody, telomerase RNA is immunoprecipitated specifically with p80 or p133 (Fig. 3B). Both antibodies also specifically immunoprecipitate p133 (Fig. 3C), which migrates at an anomalous apparent molecular mass of ≈120 kDa. We used several techniques to confirm that the protein migrating at 120 kDa is endogenous p133. Recombinant p133 made in three different expression systems migrated at 120 kDa, two independent p133 anti-peptide antibodies specifically recognized a 120-kDa Tetrahymena protein by immunoblot and immunoprecipitation, and this protein was present in telomerase-positive extracts of vegetatively grown and mated cells (data not shown). Immunoprecipitations with anti-Ct antibody, in which all activity was recovered in the bound fraction, appeared to contain roughly similar quantities of all three telomerase proteins (Fig. 3D, lane 1) when compared against equal mass amounts of recombinant standards (Fig. 3D, lanes 2–3). However, some differences in relative recovery were observed with different extracts (see Discussion). We conclude that all of the four known Tetrahymena telomerase components compose parts of the same RNP.
Nucleotide Addition Activity of Recombinant p133.
To characterize the function of p133 alone, we produced recombinant protein. To overcome the atypical codon usage in Tetrahymena, we constructed an entirely synthetic ORF encoding the p133 amino acid sequence. Because the human homolog of p133 expressed in rabbit reticulocyte lysate with human telomerase RNA is sufficient to produce apparently native-like telomerase activity in the lysate extract (13), we tested the ability of Tetrahymena p133 and telomerase RNA to do the same (Fig. 4A). Telomerase activity assays of p133 and telomerase RNA coexpressed in rabbit reticulocyte lysate under typical endogenous Tetrahymena telomerase assay conditions revealed a limited addition of dTTP and 32P-dGTP to a single-stranded, telomeric DNA primer (G4T2)3 (Fig. 4A, lane 1). Expression of p133 alone (Fig. 4A, lane 2), telomerase RNA alone (Fig. 4A, lane 3), or coexpression of telomerase RNA with p133 site-specifically substituted at the aspartic acids of motif C (Fig. 4A, lane 4) all failed to produce the nucleotide addition activity. RNase treatment of lysate containing coexpressed p133 and telomerase RNA before the telomerase assay also eliminated the nucleotide addition activity (data not shown).
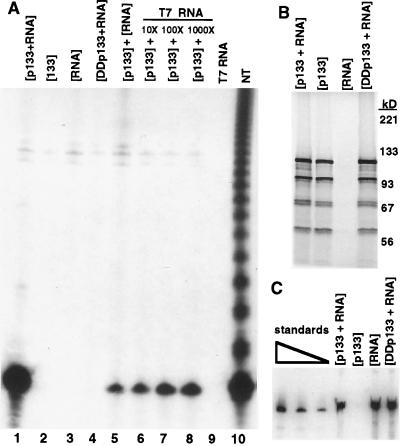
Figure 4.
Reticulocyte lysate expression of p133 and telomerase RNA. (A) Telomerase activity was assayed for p133 coexpressed with RNA [p133+RNA] (lane 1); individual components expressed alone [p133], [RNA] (lanes 2 and 3); or p133 with motif C substitution of both aspartic acids for alanines coexpressed with RNA [DDp133+RNA] (lane 4). Assays also were performed for lysate-expressed p133 and RNA mixed after synthesis (lane 5); lysate-expressed p133 mixed with purified telomerase RNA (lanes 6–8: 20 ng, 200 ng, 2 μg of RNA); purified RNA alone (lane 9: 2 μg RNA); or partially purified endogenous telomerase (NT, lane 10). The radiolabel in recombinant enzyme lanes in the top half of the gel is from lysate labeling of plasmid DNA. (B) SDS/PAGE followed by autoradiography was used to detect recombinant protein synthesized in the lysates indicated. The migration of molecular mass markers is indicated at right. (C) Northern analysis of telomerase RNA was used to detect recombinant RNA synthesized in 2.5 μl of the lysates indicated. Telomerase RNA standards were loaded at left (1.1, 0.6, 0.3 ng RNA).
When p133 and telomerase RNA were expressed separately in lysate and then combined before assaying, a reduced level of nucleotide addition activity was observed compared with the activity obtained by coexpression (Fig. 4A, compare lanes 5 and 1). Similarly, when p133 expressed in reticulocyte lysate was combined with telomerase RNA that had been purified after transcription, nucleotide addition activity was much less than that obtained by coexpression (Fig. 4A, lanes 6–8). This was true even if a vast excess of purified RNA was added to the p133 lysate. As a control, purified RNA alone had no activity in reticulocyte lysate (Fig. 4A, lane 9). Also, each lysate reaction produced comparable levels of [35S]methionine labeled p133 (Fig. 4B) and/or telomerase RNA (Fig. 4C), whether p133 and RNA were expressed together or separately. Expression of the p133 gene in lysate produced several protein species. The largest form at ≈120 kDa migrates as full length protein and cross-reacts with anti-Ct antibody, indicating that the C terminus is present. The second largest form also cross-reacts with anti-Ct antibody and thus likely contains an intact polymerase domain. Approximately 2 ng of p133 and 1 ng of telomerase RNA were produced per microliter of lysate reaction (Fig. 4C and data not shown).
In each of the assays described above, the nucleotide addition activity catalyzed by recombinant p133 and telomerase RNA extended the primer (G4T2)3 by one residue. This pattern of elongation would result from alignment of the primer 3′ end with the extreme 5′ end of the RNA template. Endogenous Tetrahymena telomerase aligns this primer 3′ end at both template 5′ and 3′ ends, adding either a single nucleotide or six before reaching the end of the template (4). Also, native Tetrahymena telomerase processively added multiple repeats to an individual primer molecule (Fig. 4A, lane 10). Dilution of native telomerase into reticulocyte lysate did not alter the processive pattern of elongation products (data not shown).
To investigate the differences between recombinant and native enzymes in primer alignment and repeat addition processivity, we assayed nucleotide addition with a variety of primers and elongation conditions. Recombinant p133+RNA extended the 18-nt primer (G4T2)3 by 1 radiolabeled dGTP (Fig. 5A, lane 1), the 18-nt primer (T2G4)3 by 3 nucleotides (lane 2), and the 19-nt G-rich but nontelomeric primer (TG)8T2G by up to 6 nucleotides (Fig. 5A, lane 3). These patterns correspond to elongation from each primer 3′ end to the end of the template, if each primer 3′ end aligned to the template with maximal perfect match. To confirm that only correctly templated sequence was added, we performed an elongation of (TG)8T2G in the presence of either 32P-dGTP, dTTP, dCTP, and dATP (Fig. 5A, lane 4) or 32P-dGTP and ddTTP (Fig. 5A, lane 5). No additional nucleotide addition was observed in the presence of all 4 dNTPs, and ddTTP prematurely terminated extension after addition of the anticipated 3 dGTP and 1 ddTTP to a primer 3′ end. Native telomerase produced similar products in the initial repeat added, extending (G4T2)3 (Fig. 5A, lane 6), (T2G4)3 (Fig. 5A, lane 7), and (TG)8T2G (Fig. 5A, lane 8) by 1, 3, and 6 nucleotides, respectively, to give the first intense DNA product. Elongation of (TG)8T2G was halted after addition of 4 nucleotides in the presence of dGTP and ddTTP (Fig. 5A, lane 9) and was not affected by the presence of all 4 dNTPs (data not shown). However, unlike recombinant p133+RNA, native telomerase added additional telomeric repeats to each substrate in a processive manner (Fig. 5A, lanes 6–8).
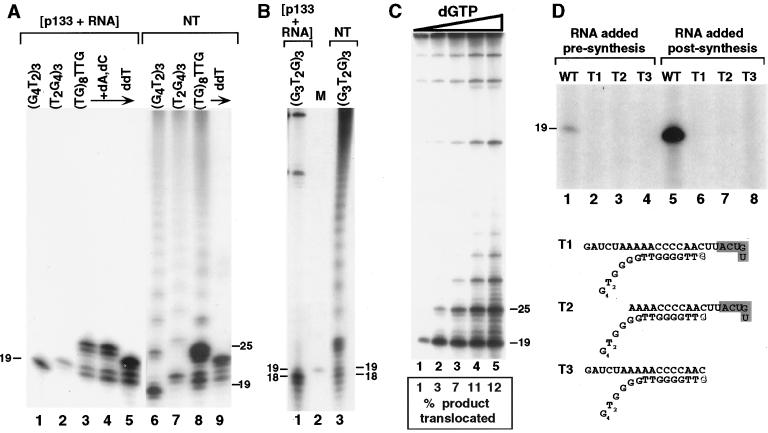
Figure 5.
Activity of recombinant p133. For each panel, numbers to the left and/or right indicate product sizes in nucleotides. (A and B) The single-stranded DNA primers indicated at top were assayed with coexpressed p133+RNA or partially purified endogenous enzyme (NT). In A, the reaction in lane 4 contained added dATP and dCTP; the reactions in lanes 5 and 9 contained ddTTP instead of dTTP. The exposures of lanes 1–5 and 6–9 were adjusted separately although all lanes were part of the same gel. In B, lane 2 is a marker (M) indicating the migration of an 18- nt telomeric primer extended by addition of one 32P-dGTP. (C) Primer (G4T2)3 was assayed with coexpressed p133+RNA and increasing concentrations of dGTP (0.6, 2, 6, 20, 60 μM) diluted 6-fold in specific activity from other reactions shown but kept at the same specific activity in the titration. The radiolabel in the top half of the gel is from lysate labeling of plasmid DNA. Note that longer product DNAs are actually less abundant than they appear because they have a higher specific activity. Below each lane, processivity at different dGTP concentrations is compared as the % of 19-nt product (+1) that translocates and is elongated to become 25-nt product (+7), normalized for the 5× greater specific activity of the longer product (i.e., the numbers are a molar percentage of +7 product relative to +1 and +7 products combined). Similar numbers were obtained by maintaining 32P-dGTP concentration and titrating unlabeled dGTP (data not shown). The stimulatory effect of dGTP appears to saturate between 10 and 100 μM. (D) The full length telomerase RNA (WT) or shorter RNAs (T1–T3) containing the template and different template-adjacent regions as shown were assayed in elongation reactions of the primer (G4T2)3. Purified RNAs (20 pmol) were either added to lysate before synthesis of p133 (lanes 1–4) or added after p133 synthesis (lanes 5–8) subsequent to addition of primer (10 pmol).
In addition to primer elongation, telomerase preparations from Tetrahymena and several other species catalyze the nucleolytic cleavage of primer or product DNA (4–6). To test for the nucleolytic cleavage activity characteristic of Tetrahymena telomerase, we assayed the products formed under standard elongation conditions with the primer substrate (G3T2G)3 (Fig. 5B). Radiolabeled products from reaction of (G3T2G)3 included primer-sized and smaller DNAs for both recombinant (Fig. 5B, lane 1) and native (Fig. 5B, lane 3) enzymes. This pattern of product DNA results from removal of one or several terminal primer nucleotides followed by replacement with new, radiolabeled synthesis. At the primer concentration used, native telomerase can align the (G3T2G)3 primer 3′ end at both ends of the template, either aligning at the template 5′ end and removing primer residues before adding radiolabeled nucleotides or aligning at the template 3′ end and adding six nucleotides before reaching the end of the template (4). Recombinant p133+RNA appears to favor alignment of (G3T2G)3 across more of the template, similar to its bias in the positioning of primer (G4T2)3. We also observed primer-sized product DNA in recombinant enzyme reactions of primer (G3T2G)3G3T2A, another native telomerase cleavage reaction substrate (ref. 4 and data not shown).
Primer elongation by native Tetrahymena telomerase is highly processive at the sub-micromolar dGTP concentration of standard assay conditions (8, 21). In contrast, for some telomerase enzymes, processive elongation within the first repeat occurs at low dGTP concentration, but processive addition of entire repeats requires much higher dGTP concentration (7, 21). Surprisingly, a titration of dGTP in assays of recombinant p133+RNA revealed that, although elevated dGTP concentration was not sufficient to restore the highly processive elongation characteristic of native Tetrahymena telomerase, it did foster a low level of processivity (Fig. 5C). This extent of dGTP-stimulated processivity in the native enzyme would be undetectable because of the much greater processivity obtained by a dGTP concentration-independent mechanism (22).
In all of the assays described above, the full length 159-nt Tetrahymena telomerase RNA was used to provide the template. For other reverse transcriptases, recognition of nucleic acid substrates involves binding and extension of a template-primer duplex. In initial assays, we tested the ability of recombinant p133+RNA to extend oligo (dG) hybridized to poly (rC) by addition of 32P-dGTP. No elongation was observed (data not shown). We next used shorter versions of the telomerase RNA itself. Three telomerase RNA templates including different lengths of template-flanking sequence were designed to form equivalent interactions with the primer (G4T2)3 (Fig. 5D, Top). In each case, addition of a single 32P-dGTP would result in elongation to the end of the template. Full length telomerase RNA and the three telomerase RNA derivatives were added to in vitro transcription/translation reactions either before synthesis of p133 (Fig. 5D, lanes 1–4) or after p133 synthesis (Fig. 5D, lanes 5–8). In either case, although the full length RNA served as template for primer elongation (Fig. 5D, lanes 1, 5), none of the smaller RNAs did (Fig. 5D, lanes 2–4, 6–8). All four templates served as substrates for the Superscript II derivative of Moloney murine leukemia virus, which actually demonstrated some preference for the shorter templates (data not shown). These results, combined with telomerase RNA mutational studies in vitro and in vivo (see ref. 9 for a recent review), suggest that the p133 active site cleft does not efficiently recognize a template-primer duplex alone.
DISCUSSION
Proteins associated with and/or required for telomerase activity have been identified by biochemical, molecular, and genetic techniques. In Tetrahymena, we show that three telomerase proteins p80, p95, and p133 and the telomerase RNA form parts of a common active RNP complex. Similarly, the human homologs of p80 and p133 have been shown to reciprocally coimmunoprecipitate (16), presumably with other as yet unidentified human telomerase proteins. Given that Tetrahymena p133 interacts with p80 and p95, why was it not obvious in the previously described chromatographic preparations of Tetrahymena telomerase (17)? Immunoblots of conventionally purified telomerase fractionated by glycerol gradient reveal that p133 is present in fractions containing the peak of telomerase activity, p80, p95, and telomerase RNA (data not shown). However, in various Tetrahymena telomerase purifications, quantitative immunoblots indicated that the amount of p133 was roughly equal to or less than telomerase RNA, whereas p80 and p95 were roughly equal to or greater than RNA (data not shown). The apparent underrepresentation of p133 in some preparations is puzzling. It may be that p133 is indeed substoichiometric. The absence of this subunit in yeast and human cells does not appear to reduce the stability of the remaining form(s) of telomerase RNP, and the sedimentation properties of the yeast RNP are not altered by the presence or absence of the TERT component in the cell (19, 23). However, estimation of p133 and RNA copy number is imprecise and could differ between endogenous and recombinant forms. Also, modification or proteolysis could diminish the amount of p133 migrating as a polypeptide of 120 kDa. In any case, there may be a previously unanticipated complexity of telomerase RNP forms, including additional subunits that remain to be identified.
When expressed in reticulocyte lysate with telomerase RNA, recombinant p133 catalyzes a reverse transcriptase activity with some similarities to and some differences from the activity of endogenous Tetrahymena telomerase. Like the native enzyme, p133+RNA can use the correct template sequence within the telomerase RNA to direct nucleotide addition and cleavage. A bias toward alignment of primers across the maximal complementary template region was observed for recombinant p133+RNA. More surprising was the limited and dGTP concentration-dependent processivity of the recombinant enzyme because no differences were reported between the activities of native human telomerase and lysate-expressed human TERT + telomerase RNA (13). It is possible that the TERT subunit and RNA from different species have dramatic differences in activity. Alternately, rabbit reticulocyte lysate components may fold, modify, or complement the two recombinant complexes differently, with the human TERT+RNA adopting a more native-like structure than the Tetrahymena TERT+RNA. Whatever the molecular explanation for the difference between recombinant and native activities described here, our results indicate that properties of telomerase mechanism such as processivity are separable from telomerase active site function.
In addition, our results indicate that features of the polymerase domain itself may be novel. The apparent inability of recombinant p133 to template DNA synthesis using typical reverse transcriptase template-primer duplexes, or even a telomeric primer annealed to shorter versions of the telomerase RNA, suggests that the telomerase active site cleft interacts with nucleic acid in a manner different from other reverse transcriptases. This idea is consistent with both in vitro and in vivo RNA mutational studies that reveal requirements for nontemplate regions of the telomerase RNA and suggest nontemplating roles for the template residues themselves (see ref. 9 for review). If nontemplate regions of the RNA interact with protein sites to appropriately position the template, the active site cleft itself need not have substantial affinity for nucleic acid. Alternately, the requirement for nontemplate regions of the telomerase RNA in the nucleotide addition reaction could be a direct consequence of their requirement for active site structure and function. Clearly, many additional models are possible and remain to be investigated biochemically.
Acknowledgments
We thank the Barker Hall DNA synthesis and sequencing facilities for excellent support, Don Rio for invaluable advice and reagents, and Michael Botchan, Nicholas Cozzarelli, Paul Kaufman, and Collins lab members for discussion and comments on the manuscript. This work was supported in part by National Institutes of Health Grant GM54198 (K.C.) and a Howard Hughes Predoctoral Fellowship (L.G.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RNP, ribonucleoprotein; TERT, telomerase reverse transcriptase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF061284).
A commentary on this article begins on page 8415.
References
- 1.Greider C W. Annu Rev Biochem. 1996;66:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 2.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 3.Yu G, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 4.Collins K, Greider C W. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 5.Cohn M, Blackburn E H. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 6.Melek M, Greene E C, Shippen D E. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greider C W, Collins K, Autexier C. In: DNA Telomerases. DePamphlis M, editor. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1996. pp. 619–638. [Google Scholar]
- 8.Greider C W. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nugent C I, Lundblad V. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 10.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 11.Gandhi L, Collins K. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 13.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichsteiner S, Kim N W, Trager J B, et al. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T, Ishikawa F. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 15.Beattie T L, Zhou W, Robinson M O, Harrington L. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 16.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S K, Mar V, Bass M B, Robinson M O. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins K, Kobayashi R, Greider C W. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 18.Autexier C, Greider C W. Genes Dev. 1994;8:563–575. doi: 10.1101/gad.8.5.563. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 20.Csank C, Taylor F M, Martindale D W. Nucleic Acids Res. 1990;18:5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond P W, Cech T R. Nucleic Acids Res. 1997;25:3698–3704. doi: 10.1093/nar/25.18.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins K, Greider C W. EMBO J. 1995;14:5422–5432. doi: 10.1002/j.1460-2075.1995.tb00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingner J, Cech T R, Hughes T R, Lundblad V. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]