Abstract
An inducible resistance determinant, ermSF, from the tylosin producer Streptomyces fradiae NRRL 2338 has been cloned, sequenced, and shown to confer inducible macrolide-lincosamide-streptogramin B resistance when transferred to Streptomyces griseofuscus NRRL 23916. From mapping studies with S1 nuclease to locate the site of transcription initiation, the ermSF message contains a 385-nucleotide 5' leader sequence upstream from the 960-nucleotide major open reading frame that encodes the resistance determinant. On the basis of the potential secondary structure that the ermSF leader can assume, a translational attenuation model similar to that for ermC is proposed. The model is supported by mutational analysis involving deletions in the proposed attenuator. By analysis with restriction endonucleases, ermSF is indistinguishable from the tlrA gene described by Birmingham et al. (V. A. Birmingham, K. L. Cox, J. L. Larson, S. E. Fishman, C. L. Hershberger, and E. T. Seno, Mol. Gen. Genet. 204:532-539, 1986) which comprises one of at least three genes from S. fradiae that can confer tylosin resistance when subcloned into S. griseofuscus. When tested for inducibility, ermSF appears to be strongly induced by erythromycin, but not by tylosin.
Full text
PDF



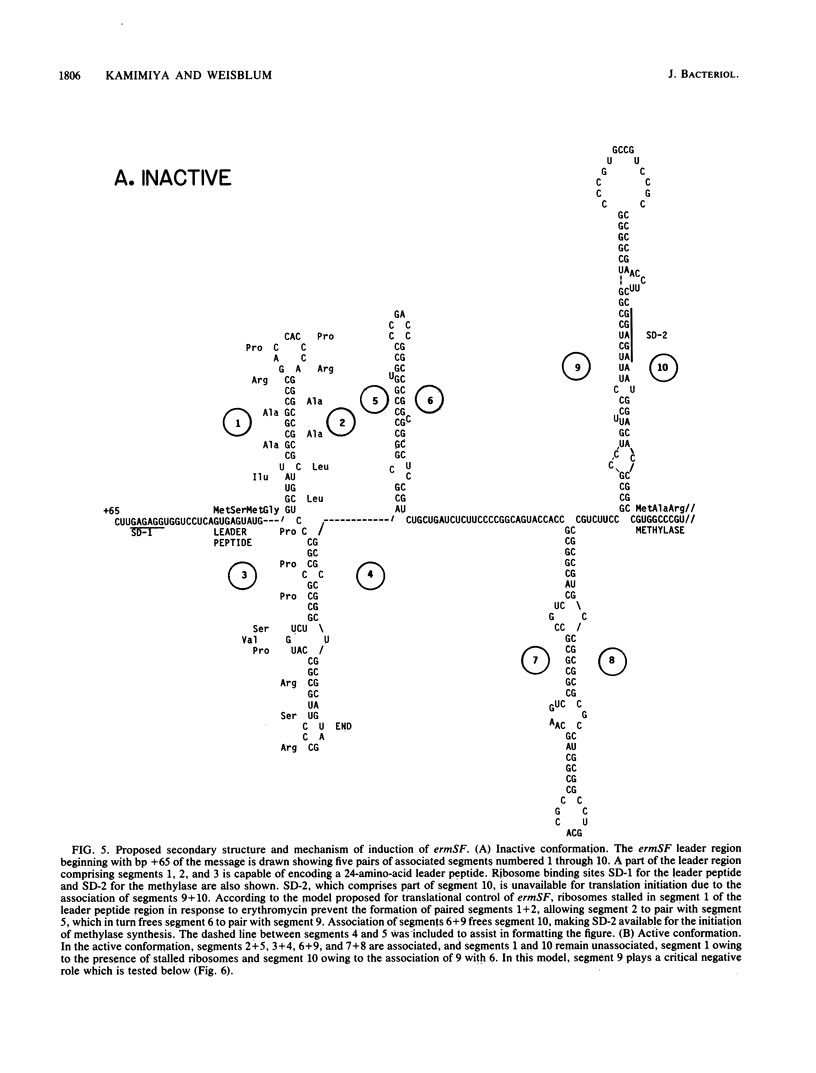
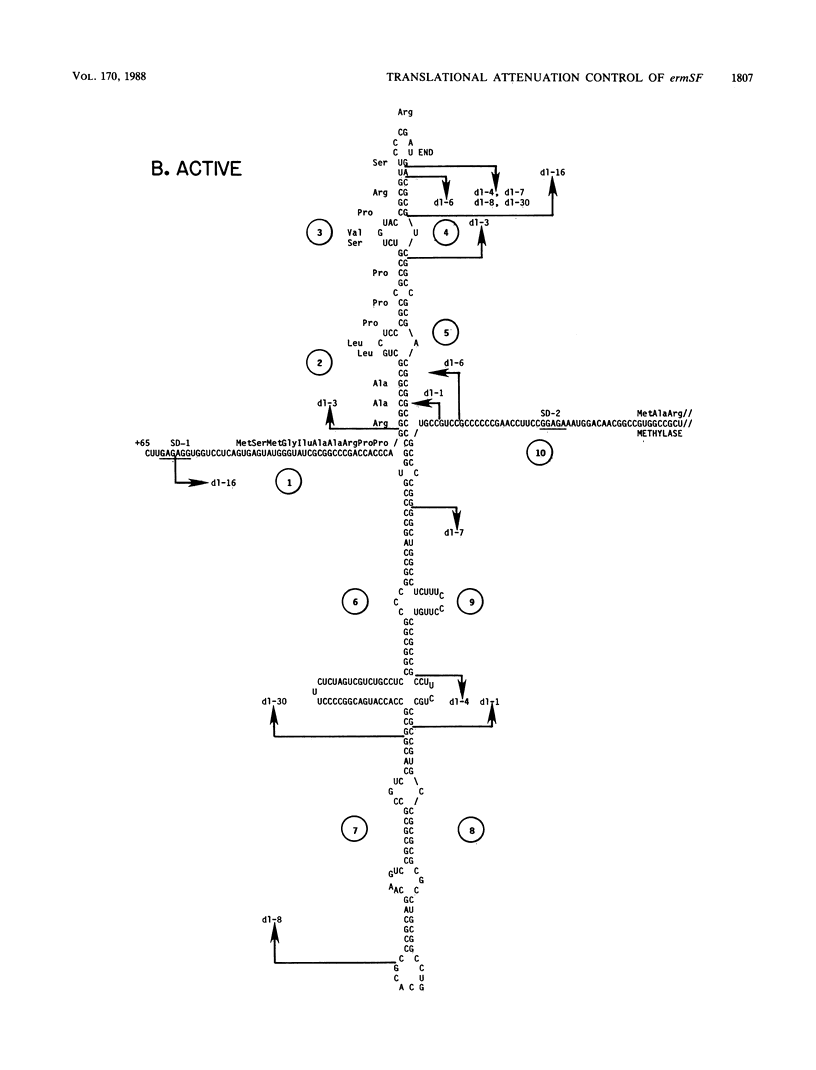

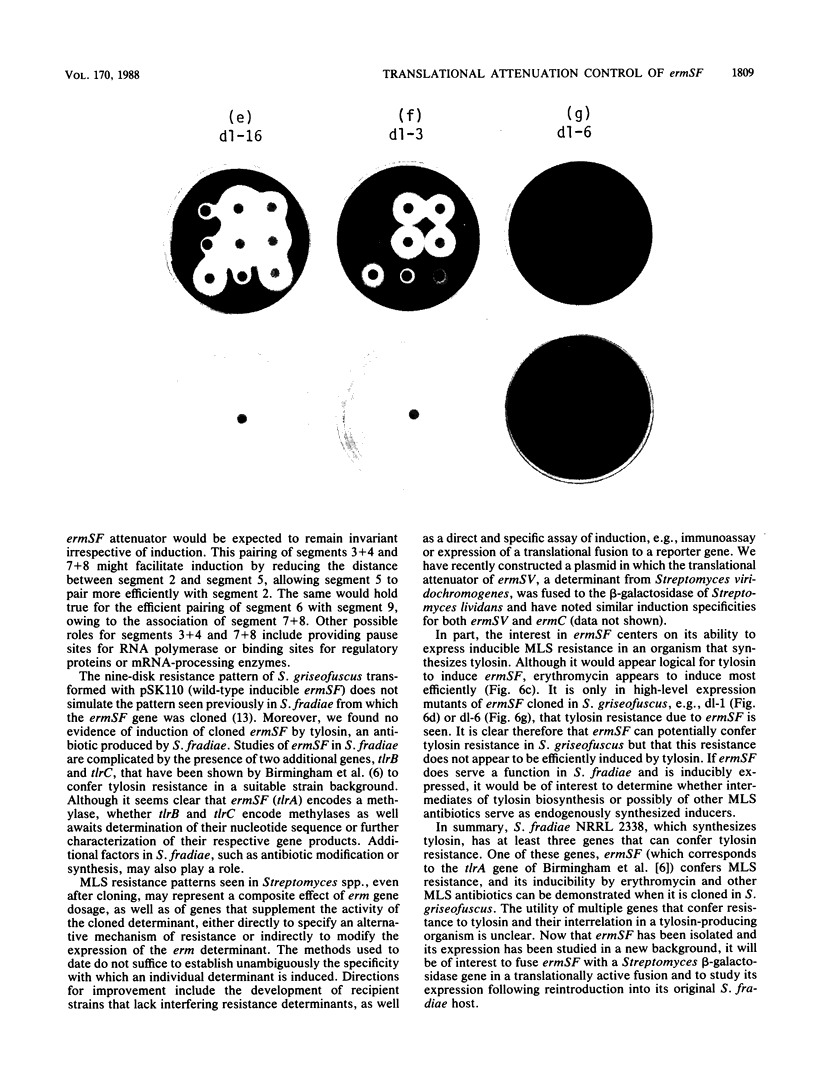


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltz R. H. Genetic recombination in Streptomyces fradiae by protoplast fusion and cell regeneration. J Gen Microbiol. 1978 Jul;107(1):93–102. doi: 10.1099/00221287-107-1-93. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Seno E. T., Stonesifer J., Wild G. M. Biosynthesis of the macrolide antibiotic tylosin. A preferred pathway from tylactone to tylosin. J Antibiot (Tokyo) 1983 Feb;36(2):131–141. doi: 10.7164/antibiotics.36.131. [DOI] [PubMed] [Google Scholar]
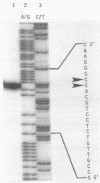
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1-3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- Birmingham V. A., Cox K. L., Larson J. L., Fishman S. E., Hershberger C. L., Seno E. T. Cloning and expression of a tylosin resistance gene from a tylosin-producing strain of Streptomyces fradiae. Mol Gen Genet. 1986 Sep;204(3):532–539. doi: 10.1007/BF00331036. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Brown N. L. Two promoters from the Streptomyces plasmid pIJ101 and their expression in Escherichia coli. Gene. 1987;51(2-3):179–186. doi: 10.1016/0378-1119(87)90306-4. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cundliffe E., Thompson J. Ribose methylation and resistance to thiostrepton. Nature. 1979 Apr 26;278(5707):859–861. doi: 10.1038/278859a0. [DOI] [PubMed] [Google Scholar]
- Docherty A., Grandi G., Grandi R., Gryczan T. J., Shivakumar A. G., Dubnau D. Naturally occurring macrolide-lincosamide-streptogramin B resistance in Bacillus licheniformis. J Bacteriol. 1981 Jan;145(1):129–137. doi: 10.1128/jb.145.1.129-137.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Epp J. K., Burgett S. G., Schoner B. E. Cloning and nucleotide sequence of a carbomycin-resistance gene from Streptomyces thermotolerans. Gene. 1987;53(1):73–83. doi: 10.1016/0378-1119(87)90094-1. [DOI] [PubMed] [Google Scholar]
- Fujisawa Y., Weisblum B. A family of r-determinants in Streptomyces spp. that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1981 May;146(2):621–631. doi: 10.1128/jb.146.2.621-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Graham M. Y., Weisblum B. 23S ribosomal ribonucleic acid of macrolide-producing streptomycetes contains methylated adenine. J Bacteriol. 1979 Mar;137(3):1464–1467. doi: 10.1128/jb.137.3.1464-1467.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T., Israeli-Reches M., Del Bue M., Dubnau D. DNA sequence and regulation of ermD, a macrolide-lincosamide-streptogramin B resistance element from Bacillus licheniformis. Mol Gen Genet. 1984;194(3):349–356. doi: 10.1007/BF00425543. [DOI] [PubMed] [Google Scholar]
- Hahn J., Grandi G., Gryczan T. J., Dubnau D. Translational attenuation of ermC: a deletion analysis. Mol Gen Genet. 1982;186(2):204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Byeon W. H., Weisblum B. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983 Jun;154(3):1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. Shuttle vectors for cloning recombinant DNA in Escherichia coli and Streptomyces griseofuscus C581. J Bacteriol. 1984 Jan;157(1):314–317. doi: 10.1128/jb.157.1.314-317.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Maas R. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid. 1983 Nov;10(3):296–298. doi: 10.1016/0147-619x(83)90045-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Monod M., Mohan S., Dubnau D. Cloning and analysis of ermG, a new macrolide-lincosamide-streptogramin B resistance element from Bacillus sphaericus. J Bacteriol. 1987 Jan;169(1):340–350. doi: 10.1128/jb.169.1.340-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol. 1985 May;162(2):633–640. doi: 10.1128/jb.162.2.633-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattee P. A. Use of tetrazolium for improved resolution of bacteriophage plaques. J Bacteriol. 1966 Sep;92(3):787–788. doi: 10.1128/jb.92.3.787-788.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts A. N., Hudson G. S., Brenner S. An erythromycin-resistance gene from an erythromycin-producing strain of Arthrobacter sp. Gene. 1985;35(3):259–270. doi: 10.1016/0378-1119(85)90004-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982 Nov;20(1):51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]
- Uchiyama H., Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38(1-3):103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Siddhikol C., Lai C. J., Demohn V. Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J Bacteriol. 1971 Jun;106(3):835–847. doi: 10.1128/jb.106.3.835-847.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]