Abstract
The Na+/H+ exchanger regulatory factor (NHERF) binds to the tail of the β2-adrenergic receptor and plays a role in adrenergic regulation of Na+/H+ exchange. NHERF contains two PDZ domains, the first of which is required for its interaction with the β2 receptor. Mutagenesis studies of the β2 receptor tail revealed that the optimal C-terminal motif for binding to the first PDZ domain of NHERF is D-S/T-x-L, a motif distinct from those recognized by other PDZ domains. The first PDZ domain of NHERF-2, a protein that is 52% identical to NHERF and also known as E3KARP, SIP-1, and TKA-1, exhibits binding preferences very similar to those of the first PDZ domain of NHERF. The delineation of the preferred binding motif for the first PDZ domain of the NHERF family of proteins allows for predictions for other proteins that may interact with NHERF or NHERF-2. For example, as would be predicted from the β2 receptor tail mutagenesis studies, NHERF binds to the tail of the purinergic P2Y1 receptor, a seven-transmembrane receptor with an intracellular C-terminal tail ending in D-T-S-L. NHERF also binds to the tail of the cystic fibrosis transmembrane conductance regulator, which ends in D-T-R-L. Because the preferred binding motif of the first PDZ domain of the NHERF family of proteins is found at the C termini of a variety of intracellular proteins, NHERF and NHERF-2 may be multifunctional adaptor proteins involved in many previously unsuspected aspects of intracellular signaling.
PDZ domains are conserved protein modules that mediate protein–protein interactions (1–3). The term “PDZ” is derived from the first letters in the names of the three proteins in which these modules were originally characterized: PSD-95, Dlg, and ZO-1. PDZ domains bind to the C-terminal tails of target proteins, and the binding preferences of a number of PDZ domains have been characterized (4–7). Such characterization is useful because it allows for predictions regarding the set of proteins to which a given PDZ domain may potentially bind.
A PDZ domain-containing protein, the Na+/H+ exchanger regulatory factor (NHERF), has been recently characterized (8) and found to regulate the Na+/H+ exchanger type 3 (NHE3) (8–9) via an interaction which does not involve binding of the NHE3 C terminus to the NHERF PDZ domains (9). The function and preferences of the two NHERF PDZ domains were unknown until NHERF was identified as a binding partner of the β2-adrenergic receptor (10). The interaction of NHERF with the β2 receptor is mediated via binding of the first PDZ domain of NHERF to the last few amino acids of the β2 receptor tail (10). In the present study, we characterize this interaction via mutagenesis of the β2 receptor tail and saturation NHERF-binding studies. We furthermore demonstrate that NHERF-2, a close relative of NHERF, specifically binds to the β2 receptor tail and exhibits binding specificity similar to NHERF. Moreover, we demonstrate how the binding preferences of the first PDZ domain of the NHERF family of proteins account for previously reported interactions involving NHERF-2 and allow for predictions regarding other potential NHERF- and NHERF-2-binding partners, such as the purinergic P2Y1 receptor and cystic fibrosis transmembrane conductance regulator (CFTR).
MATERIALS AND METHODS
NHERF PDZ1 Overlays.
Hexahistidine- and S-tagged NHERF domain 1 fusion protein (“PDZ1”; residues 1–151 of full-length NHERF) was created via insertion of rabbit NHERF cDNA (8) into pET-30A (Novagen) and expression according to manufacturer’s instructions. Wild-type or mutant human β2 receptor tails (10), as well as human P2Y1 and P2Y2 receptor tails, were expressed as glutathione S-transferase (GST) fusion proteins and examined for NHERF binding via a far Western blot overlay technique. Mutant β2 receptor tail GST constructs were created by PCR amplification from native receptor cDNA’s by using mutant sequence oligonucleotides and inserting the PCR products into the pGEX-2T vector (Pharmacia); all mutations were confirmed via sequencing with an Applied Biosystems 377 automated sequencer. The GST fusion proteins (25 μg/lane) were run on 4–20% SDS/PAGE gels (NOVEX, San Diego), blotted, and overlaid with increasing concentrations of NHERF PDZ1 fusion protein in 2% milk and 0.1% Tween-20 in PBS (“blot buffer”) for 1 hr at room temperature. The blots were then washed three times with blot buffer, incubated for 1 hr at room temperature with an horseradish peroxidase-conjugated anti-S-tag antibody (Novagen) in blot buffer, washed three more times with blot buffer, and visualized via chemiluminescence. To perform saturation-binding curves, equal amounts of a given fusion protein were loaded into multiple lanes, and the resultant blots were cut into one-lane strips. The strips were incubated with increasing concentrations of NHERF PDZ1 fusion protein, and the amount of NHERF binding for each was quantified via scanning and analysis with the program iplab (Signal Analytics).
Cloning of NHERF-2.
NHERF-2 was cloned from a human lung cDNA library (CLONTECH) by amplification using primers (5′-CACCCGGAATTCGCCGCCATGGC-CGCGCCGGAGCCGCTGCGG and 5′-CACCCGGTCGACTCAGTCAGCAGGCC-CCGGCAGCGA) based on the sequence for “TKA-1,” which was submitted to GenBank by Seedorf and Ullrich (GenBank accession no. Z50150; unpublished results). The species we cloned (GenBank accession no. AF035771) is clearly the same gene product as TKA-1, E3KARP (9), and SIP-1 (11), although none of these three sequences is identical to any other and our sequence is slightly different from each of them. We prepared full-length NHERF-2 as well as the two halves of the molecule as fusion proteins by inserting amplified fragments into pET-30A by using EcoRI and SalI (underlined sites in primers); the “N-terminal” NHERF-2 construct contains the first 148 amino acids and the first PDZ domain, whereas the “C-terminal” construct contains the last 189 residues and the second PDZ domain.
β2 Receptor Tail-GST Overlays.
Far Western overlay experiments were performed by running the NHERF-2 fusion protein samples (25 μg per lane) on 4–20% SDS/PAGE gels (Novex) for 1 hr at 150 V and then blotting to nitrocellulose for 40 min at 12 V. The blots were blocked in blot buffer and incubated with the wild-type β2 receptor tail expressed as a GST fusion protein (500 nM) in blot buffer for 1 hr at room temperature. The blots were washed three times with blot buffer and then incubated with an anti-GST mAb (Santa Cruz) for l hr at room temperature to detect the overlaid β2 receptor tail. The blots were then washed three more times with blot buffer and incubated for l hr at room temperature with an anti-mouse horseradish peroxidase-coupled secondary antibody (Amersham) in blot buffer. The blots were then washed three more times with blot buffer, once with PBS, and then developed with an enhanced chemiluminescence kit (Amersham) and exposed to Kodak X-Omat film for varying lengths of time.
Overlays with 35S in Vitro-Translated Protein.
The following proteins were in vitro translated by using the TNT system (Promega) in the presence of [35S]methionine: full-length CFTR-4, CFTR-K1468X, or fusion proteins of CFTR cloned into the pET33b vector (Novagen) (C-terminal amino acids 1387–1480 or N-terminal amino acids 1–80). Immobilized full-length NHERF was incubated overnight with each of the in vitro translates (2 μl of reticulocyte lysate in 10 ml of blot buffer), washed with blot buffer, dried, and exposed to film.
Overlays of Airway Protein.
Human airway epithelial cells were cultured at the air-liquid interface as described (12). Triton X-100 soluble and insoluble proteins were prepared as described (13), electrophoresed on 6–18% SDS/PAGE, and either stained with Coomassie blue or transferred to Immobilon. Immobilized proteins were blocked with blot buffer, incubated with anti-NHERF antibody (1:1000 in blot buffer), washed three times in blot buffer, and then incubated with anti-rabbit horseradish peroxidase secondary antibody (Amersham) (1:2500). Reactive proteins were visualized with Super-Signal (Pierce). Identical lanes were incubated with a 32P-phosphorylated probe of the C terminus of CFTR cloned into the pET33b vector, which contains a PKA-phosphorylation site. The protein was expressed in BL21 (DE3) cells, purified on a Talon metal affinity column (Pharmacia), phosphorylated with PKA and 32P-ATP, and dialyzed to remove nonincorporated ATP. The membrane was washed three times with blot buffer, dried, and exposed to film.
Estimation of NHERF Cellular Concentration.
Various dilutions of full-length NHERF fusion protein were run alongside samples of crude kidney extract on SDS/PAGE. The samples were blotted and the amount of NHERF in each sample was visualized by Western blotting using a previously described anti-NHERF antibody (10) and a horseradish peroxidase-coupled anti-rabbit Ig secondary antibody (Amersham). The resultant bands were scanned and quantified, and the amount of NHERF in the crude tissue was derived from the fusion protein standard curves. The per unit protein concentration of NHERF was translated into a molar concentration by assuming that 30% of the weight of the average human kidney (150 g) is protein and that one-half of the volume of the average human kidney (216 ml) is intracellular volume.
RESULTS
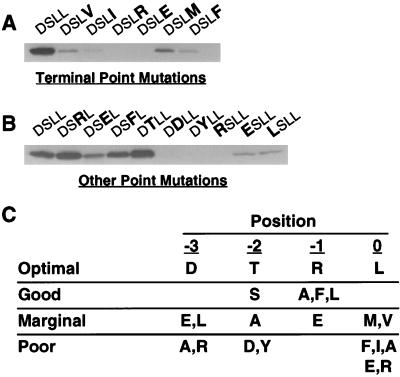
The binding preferences of the first PDZ domain of NHERF were determined via mutagenesis of the β2 receptor tail, the last four amino acids of which are D-S-L-L. Previous work demonstrated that mutation of the terminal leucine of the β2 receptor tail to alanine completely abolishes binding of NHERF (10). Mutation of this position to six other amino acids (Fig. 1A) revealed that the wild-type leucine is strongly preferred. Even amino acids with similar side chains such as isoleucine and valine yield more than fivefold reductions in NHERF binding when they are substituted for leucine at the end of the β2 receptor tail.
Figure 1.
NHERF preferentially binds to the motif D-S/T-x-L at the end of target proteins. (A) Binding of the first PDZ domain of NHERF to the β2 receptor tail is inhibited by point mutations of the final residue of the tail. GST fusion proteins of the β2 receptor tail with mutations at the terminal position were expressed and purified. Equal amounts (≈25 μg) were loaded on SDS/PAGE gels, blotted, and overlaid with NHERF PDZ1 fusion protein (50 nM). The mutations of the β2 receptor tail (D-S-L-L) are indicated in bold at the top of the panel. (B) Point mutations at the −1, −2, and −3 positions of the β2 receptor tail differentially alter NHERF PDZ1 binding. Equal amounts of various β2 receptor tail point mutants were loaded on an SDS/PAGE gel, blotted, and probed for NHERF PDZ1 fusion protein binding. (C) Summary of NHERF PDZ1-binding preferences at the final four amino acid positions of target proteins based on single amino acid substitution studies of the β2 receptor tail. The optimal amino acid for each position is shown in the first line of the table. “Good” means that the given change causes less than a twofold loss in NHERF PDZ1 binding relative to the optimal amino acid at that position, whereas “marginal” means that the given change causes more than a twofold but less than a 10-fold decrease in binding. “Poor” means that the given change leads to a >10-fold decrease in NHERF PDZ1 binding. The results are representative of two to four independent determinations for binding to each mutant tail.
Previous studies revealed that mutation to alanine at the −2 or −3 positions of the β2 receptor tail significantly inhibits NHERF binding but that mutation to alanine at the −1 position has no effect (10). Further mutations at the −1, −2, and −3 positions (Fig. 1B) revealed that aspartate is markedly better than any other amino acid examined at the −3 position but that threonine is slightly better than serine at the −2 position. There is a slight preference at the −1 position for basic residues over neutral or acidic residues (arginine is better than leucine, and leucine is better than glutamate), but the difference is less than a twofold change in NHERF binding either way compared with the wild-type β2 receptor tail. Mutation to aspartate at the −2 position results in a complete loss of NHERF binding, suggesting that phosphorylation of this site in vivo is likely to inhibit the binding of NHERF to the β2 receptor. Interestingly, this serine (Ser411) is a substrate for phosphorylation by the G protein-coupled receptor kinase GRK5 (14). The effects of point mutations at each of the final four positions of the β2 receptor tail are summarized in Fig. 1C.
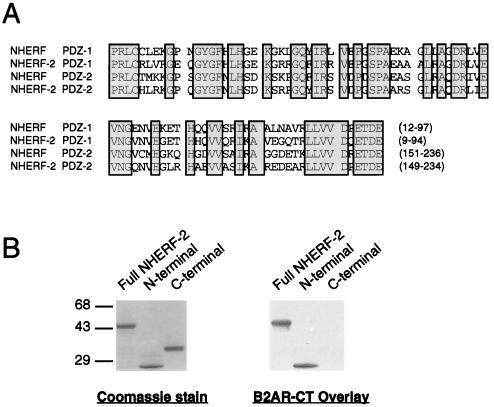
Using primers based on GenBank accession no. Z50150, a close relative of NHERF was cloned from a human lung cDNA library. This species will be referred to here as “NHERF-2.” The two PDZ domains of NHERF-2, like the two PDZ domains of NHERF, exhibit ≈60% identity with each other and 20–30% identity with the PDZ domains of other proteins such as PSD-95 and Dlg. In place of the G-L-G-F sequence found at the core of the earliest characterized PDZ domains, the NHERF PDZ domains contain the sequence G-Y-G-F. An alignment of the NHERF and NHERF-2 PDZ domains is shown in Fig. 2A. Analysis of the binding of the β2 receptor tail to NHERF-2 revealed that the receptor tail binds to full-length NHERF-2 and to the first NHERF-2 PDZ domain but not to the second PDZ domain (Fig. 2B). These findings are identical to the situation for β2 receptor tail binding to NHERF (10). NHERF and NHERF-2 also exhibit a very similar pattern of preferences for the panel of β2 receptor tail mutants presented in Fig. 1 (data not shown).
Figure 2.
NHERF-2 exhibits structural and functional similarity to NHERF. (A) Alignment of NHERF and NHERF-2 PDZ domains. The first (“PDZ-1”) and second (“PDZ-2”) PDZ domains of both human NHERF and human NHERF-2 are aligned, with conserved residues boxed and shaded. The positions of the PDZ domains within the NHERF proteins are indicated by the numbers in parentheses at the end of each sequence. The two PDZ-1 sequences exhibit 73% identity, whereas the two PDZ-2 sequences exhibit 70% identity. The PDZ-1 and PDZ-2 domains of each protein are 60% identical to each other. (B) The β2 receptor tail binds to recombinant NHERF-2 fusion proteins on a blot overlay. The β2 receptor tail expressed as a GST fusion protein was overlaid onto three blotted samples: full-length NHERF-2 (“full”), the N-terminal 121 amino acids of NHERF-2, which contains the first PDZ domain (“N-term”), and the C-terminal 210 amino acids of NHERF-2, which contains the second PDZ domain (“C-term”). As shown by the Coomassie-stained SDS/PAGE gel (Left), equal amounts of the three fusion proteins were loaded. Overlays of the blotted samples with the β2 receptor tail-GST (Right) revealed that the receptor tail binds specifically to full-length NHERF-2 and to the N-terminal portion of NHERF-2 but not to the C-terminal portion. This experiment was performed three times with identical results.
Database searches for other proteins containing consensus C-terminal sequences appropriate for interaction with the first PDZ domain of the NHERF proteins yield a number of intracellular proteins. Some of these proteins are listed in Table 1. The list of potential NHERF-binding partners includes several proteins that are the products of known human disease genes, such as CFTR and the Menke’s disease gene protein (MNK). The list also includes a number of transmembrane receptors. In addition to the β2-adrenergic receptor, which is known to bind to NHERF in an agonist-dependent fashion in whole cells (10), the platelet-derived growth factor receptor and the P2Y1-purinergic receptor represent potential cellular-binding partners for the NHERF proteins.
Table 1.
Potential NHERF-binding proteins
| Receptors | C-terminal motif |
| Beta-2 adrenergic receptor | D-S-L-L |
| Purinergic P2Y1 receptor | D-T-S-L |
| Platelet-derived growth factor receptor | D-S-F-L |
| Transporters | |
| Cystic fibrosis transmembrane regulator | D-T-R-L |
| Copper transporter (Menke’s disease gene) | D-T-A-L |
| Endomembrane proton pump, 58 kDa subunit | D-T-A-L |
| Other Proteins | |
| Phospholipase C-beta 1 | D-T-P-L |
| Podocalyxin | D-T-H-L |
| Kunjin virus-specified protein | D-T-V-L |
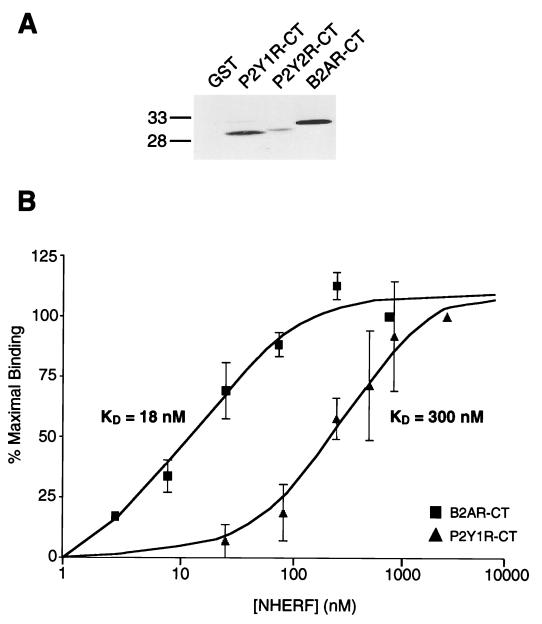
The idea that binding partners for the NHERF proteins may be identified based on the presence of the C-terminal D-S/T-x-L motif was tested by preparing the C-terminal tails of the P2Y1 and P2Y2 receptors as GST fusion proteins and examining their ability to bind NHERF in the overlay assay. These two purinergic receptor subtypes exhibit 33% overall sequence identity but are divergent at their C termini: the P2Y1 receptor ends in a good motif for NHERF PDZ1 binding (D-T-S-L) whereas the P2Y2 receptor ends in a motif that is less than ideal owing to the lack of a hydroxyl-bearing amino acid at the −2 position (D-I-R-L). As shown in Fig. 3A, the P2Y1 receptor tail exhibits robust binding of NHERF, whereas the P2Y2 receptor tail exhibits poor NHERF binding. The affinity of NHERF PDZ1 for the P2Y1 receptor tail was quantified via saturation-binding studies and compared with the NHERF-binding affinity of the β2 receptor tail (Fig. 3B); the KD for β2 receptor tail binding to NHERF PDZ1 is 18 nM, whereas the KD for P2Y1 receptor tail binding is 300 nM.
Figure 3.
The P2Y1 receptor tail binds to the first PDZ domain of NHERF, though with lower affinity than the β2-adrenergic receptor tail. (A) The first PDZ domain of NHERF binds well to the β2 receptor tail, moderately to the P2Y1 receptor tail, and poorly to the P2Y2 receptor tail. Equal amounts of the three fusion proteins (25 μg), along with control GST (Left lane), were run on an SDS/PAGE gel, blotted, and overlaid with NHERF PDZ1 fusion protein (50 nM). This experiment was performed twice with identical results. (B) Determination of the binding affinity of NHERF for the β2 receptor and P2Y1 receptor tails. Equal amounts of β2 receptor tail GST fusion protein or P2Y1 receptor tail GST fusion protein (25 μg) were run on SDS/PAGE gels, blotted, and overlaid with six different concentrations of purified S-tagged NHERF fusion protein. The x-axis is plotted as a logarithmic scale of the concentration of NHERF PDZ1 fusion protein, and the y-axis represents the relative amount of NHERF PDZ1 binding expressed as a percentage of the binding observed at the highest NHERF PDZ1 concentration (1 μM in the case of the β2 receptor, 3 μM in the case of the P2Y1 receptor). The estimated KD for NHERF binding to the β2 receptor tail is 18 nM, whereas the estimate for NHERF binding to the P2Y1 receptor tail is 300 nM. Each point and error bar represents the mean ± SEM for three independent determinations.
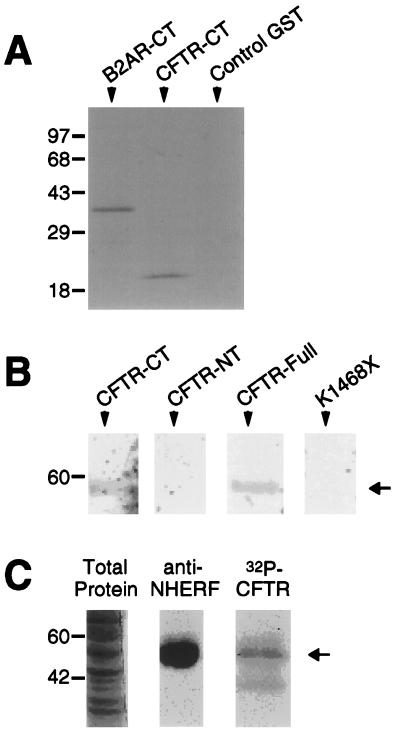
The hypothesis that interaction with the NHERF family of proteins may be predicted from the presence of a C-terminal D-S/T-x-L motif was further tested by examining the binding of NHERF to the tail of CFTR, which ends in D-T-R-L. Blotted CFTR tail fusion protein exhibited robust and specific binding of overlaid NHERF PDZ1 (Fig. 4A). The ability of full-length NHERF to interact with full-length CFTR was tested by in vitro translating full-length CFTR in the presence of [35S]methionine and overlaying it onto blotted full-length NHERF (Fig. 4B). Full-length CFTR bound avidly to NHERF, as did the C-terminal tail of CFTR when assayed separately. Conversely, neither the N-terminal portion of CFTR nor a C-terminally truncated mutant CFTR (K1468X) exhibited detectable binding to NHERF. CFTR tail bound not only to recombinantly expressed NHERF but also to native NHERF from differentiated airway epithelial cells. As shown in Fig. 4C, a Triton X-100 soluble fraction derived from cultured airway epithelia revealed a band of appropriate molecular mass (50 kDa) when probed with either an anti-NHERF antibody (10) or with 32P-labeled CFTR C-terminal fusion protein.
Figure 4.
NHERF associates with CFTR tail and full-length CFTR. (A) NHERF PDZ1 binds equally well to the tails of the β2 receptor and CFTR expressed as fusion proteins but does not bind detectably to control GST. Approximately 25 μg of each protein was run on an SDS/PAGE gel, blotted, and then overlaid with 50 nM NHERF PDZ1 fusion protein. This experiment was performed twice with identical results. (B) Full-length CFTR binds to full-length NHERF. In vitro translated 35S-labeled probes of either CFTR C terminus (“CFTR-CT”), CFTR N terminus (“CFTR-NT”), full-length CFTR (“CFTR-Full), or a truncated CFTR (”K1468X“) were overlaid onto blot strips containing ≈10 μg of purified full-length NHERF fusion protein. Full-length CFTR and CFTR C terminus bound avidly to the NHERF but CFTR N terminus and CFTR K1468X did not detectably bind. (C) An airway epithelial protein consistent with the size of NHERF is recognized by the anti-NHERF antibody and by the C terminus of CFTR. Soluble fractions of differentiated airway cells were stained with Coomassie blue and probed with an anti-NHERF antibody or with a 32P-phosphorylated C-terminal fusion protein of CFTR. The antibody and the C-terminal probe both recognized a band of identical size in the soluble fraction of airway proteins consistent with the expected size of NHERF.
DISCUSSION
The results presented here demonstrate that the first PDZ domain of NHERF family proteins binds to target proteins which terminate in the C-terminal motif D-S/T-x-L. The first PDZ protein for which a preferred C-terminal motif was identified was PSD-95, which recognizes the motif T/S-x-V at the C terminus of target proteins (4). The motif recognized by NHERF is distinct in that leucine is strongly preferred over valine at the terminal position and aspartate is preferred at the −3 position. Screening of peptide libraries has helped to define the binding partner preferences of a number of PDZ domains (5), and at the present time, no other PDZ domain has been reported to have exactly the same preferences as the first PDZ domain of the NHERF proteins. Thus, it is likely that the first NHERF PDZ domain binds to a unique set of intracellular partners that does not significantly overlap with the sets of proteins bound by other PDZ domains.
The nomenclature of the NHERF family of proteins is currently somewhat confusing. NHERF was originally purified and cloned from rabbit tissue (8). Human NHERF was recently cloned (15–16) and termed “hNHERF” (human NHERF) (16) or “ERM-binding protein 50” (15) because it was found to bind to the ERM family of actin-binding proteins (ezrin, radixin, and moesin). NHERF-2, the close relative of NHERF reported in the present manuscript, also has been cloned recently by three independent groups who have given the protein three different names: “TKA-1” (Seedorf and Ullrich, unpublished data; GenBank accession no. Z50150), “E3KARP” (9), and “SIP-1” (11). To simplify the nomenclature in this area, we propose to call this species NHERF-2 because it is closely related to NHERF (52% sequence identity) and has been shown to regulate NHE3 (9). NHERF-2 (GenBank accession no. AF035771) is essentially identical to TKA-1 and E3KARP and is also identical to SIP-1 except for a 33-bp insertion that is present in NHERF-2 but not in SIP-1. It is likely that NHERF-2 and SIP-1 represent alternatively spliced variants of the same gene.
The finding that NHERF and NHERF-2 both recognize the motif D-S/T-x-L at the C terminus of target proteins is of interest because NHERF-2 has been identified as a binding partner for two proteins which contain variants of the motif: the platelet-derived growth factor (PDGF) receptor (Seedorf and Ullrich, unpublished data; GenBank accession no. Z50150) and the testis-determining factor SRY (11). The last four amino acids of human PDGF-A and PDGF-B receptors are D-S-F-L, consistent with the C-terminal motif determined in the present study to be optimal for NHERF PDZ1 binding. Similarly, human SRY terminates in W-T-K-L, a C-terminal motif that would be predicted from our mutagenesis studies to bind NHERF-family proteins with a reasonable affinity.
The work described in the present report not only explains the interaction of NHERF family proteins with two previously identified binding partners, it also identifies two new potential cellular NHERF-associated proteins: the P2Y1 receptor and CFTR. Although the affinity of NHERF for the tail of the P2Y1 receptor is >10-fold lower than its affinity for the tail of the β2 receptor, this interaction could still potentially be of physiological relevance given the abundance of NHERF in some cell types. We have performed quantitative Western-blotting studies, which indicate that NHERF represents 0.005–0.01% of the total protein in crude kidney tissue (data not shown). This result corresponds to a likely average intracellular NHERF concentration of 500 nM to 1 μM (see Materials and Methods for an explanation of the calculations behind these estimates). NHERF may thus potentially bind to the P2Y1 receptor in a manner that is of physiological interest, and such an interaction may have relevance for recent observations that agonist stimulation of P2Y1 receptors can lead in some instances to physiological responses that are independent of G protein activation (17).
The interaction of NHERF with CFTR also may be of physiological interest. Mutations in CFTR cause cystic fibrosis (18). CFTR is a chloride channel, but some cellular actions of CFTR are believed to be independent of its channel activity (19–22). NHERF joins syntaxin (23) on a short list of CFTR-interacting proteins that might potentially mediate some of these channel-independent actions of CFTR. Interestingly, a naturally occurring truncation of the tail of CFTR (S1455X) has been recently reported (24). This mutation deletes the NHERF-binding motif characterized in the present study and produces an interesting clinical phenotype, with subjects exhibiting elevated sweat chloride concentrations but no other manifestations of cystic fibrosis. These findings suggest that the binding of NHERF or NHERF-related proteins to the tail of CFTR may be important for some aspects of CFTR function.
In summary, the motif D-S/T-x-L at the end of the β2 receptor is necessary for high affinity interaction with the first PDZ domain of the NHERF proteins. The presence of this motif at the C terminus of proteins is a useful indicator of potential NHERF binding, as we have shown here in verifying that the C termini of the P2Y1 receptor and CFTR bind to NHERF. Consensus NHERF-binding motifs are found at the C termini of many intracellular proteins. The variety of proteins possessing C termini appropriate for high-affinity binding to NHERF and NHERF-2 suggests that the NHERF proteins may be multifunctional adaptor proteins involved in many diverse aspects of cellular signaling.
Acknowledgments
We thank Shirish Shenolikar and Ed Weinman for providing NHERF cDNA constructs and advice, Rob Nicholas for P2Y1 and P2Y2 cDNA constructs, Julie Pitcher, Bob Spurney and Joe Cotten for helpful discussions regarding this work, Millie McAdams and Judy Phelps for sequencing, and Donna Addison and Mary Holben for help in the preparation of this manuscript. This work was supported in part by Grant HL16037 from the National Institutes of Health to R.J.L. and Grants HL29851 and HL42385 from the National Institutes of Health to M.J.W.
ABBREVIATIONS
- NHERF
Na+/H+ exchanger regulatory factor
- CFTR
cystic fibrosis transmembrane conductance regulator
- GST
glutathione S-transferase
Note Added in Proof
While this manuscript was under review, Wang et al. (25) reported an affinity selection characterization of NHERF PDZ domain-binding preferences and the interaction of NHERF with CFTR.
Footnotes
Data deposition: The sequence for NHERF-2 reported in this manuscript has been deposited in the GenBank database (accession no. AF035771).
References
- 1.Sheng M. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 2.Harrison S C. Cell. 1996;86:341–343. doi: 10.1016/s0092-8674(00)80105-1. [DOI] [PubMed] [Google Scholar]
- 3.Saras J, Heldin C-H. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 4.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 5.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 6.Schepens J, Cuppen E, Wieringa B, Hendriks W. FEBS Lett. 1997;409:53–56. doi: 10.1016/s0014-5793(97)00481-x. [DOI] [PubMed] [Google Scholar]
- 7.Saras J, Engstrom U, Gonez L J, Heldin C-H. J Biol Chem. 1997;272:20979–20981. doi: 10.1074/jbc.272.34.20979. [DOI] [PubMed] [Google Scholar]
- 8.Weinman E J, Steplock D, Wang Y, Shenolikar S. J Clin Invest. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun C H C, Oh S, Zizak M, Steplock D, Tsao S, Tse C-M, Weinman E J, Donowitz M. Proc Natl Acad Sci USA. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall R A, Premont R T, Chow C-W, Blitzer J T, Pitcher J A, Claing A, Stoffel R H, Barak L S, Shenolikar S, Weinman E J, et al. Nature (London) 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 11.Poulat F, de Santa Barbara P, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P. J Biol Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- 12.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 13.Hinck L, Nathke I S, Papkoff J, Nelson W J. J Cell Biol. 1994;125:1327–1340. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredericks Z L, Pitcher J A, Lefkowitz R J. J Biol Chem. 1996;271:13796–13803. doi: 10.1074/jbc.271.23.13796. [DOI] [PubMed] [Google Scholar]
- 15.Reczek D, Berryman M, Bretscher A. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 17.O’Grady S M, Elmquist E, Filtz T M, Nicholas R A, Harden T K. J Biol Chem. 1996;271:29080–29087. doi: 10.1074/jbc.271.46.29080. [DOI] [PubMed] [Google Scholar]
- 18.Welsh M J, Smith A E. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 19.Stutts M J, Canessa C M, Olsen J C, Hamrick M, Cohn J A, Rossier B C, Boucher R C. Science. 1995;269:847–850. doi: 10.1126/science.7543698. [DOI] [PubMed] [Google Scholar]
- 20.Schwiebert E M, Egan M, Hwang T-H, Fulmer S B, Allen S S, Cutting G R, Guggino W B. Cell. 1995;81:1063–1073. doi: 10.1016/s0092-8674(05)80011-x. [DOI] [PubMed] [Google Scholar]
- 21.McNicholas C M, Guggino W B, Schwiebert E M, Hebert S C, Giebisch G, Egan M E. Proc Natl Acad Sci USA. 1996;93:8083–8088. doi: 10.1073/pnas.93.15.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stutts M J, Rossier B C, Boucher R C. J Biol Chem. 1997;272:14037–14040. doi: 10.1074/jbc.272.22.14037. [DOI] [PubMed] [Google Scholar]
- 23.Naren A P, Nelson D J, Xie W, Jovov B, Pevsner J, Bennett M K, Benos D J, Quick M W, Kirk K L. Nature (London) 1997;390:302–305. doi: 10.1038/36882. [DOI] [PubMed] [Google Scholar]
- 24.Mickle J E, Macek M, Jr, Fulmer-Smentek S B, Egan M M, Schwiebert E, Guggino W, Moss R, Cutting G R. Hum Mol Genet. 1998;7:729–735. doi: 10.1093/hmg/7.4.729. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Raab R W, Schatz P J, Guggino W B, Li M. FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]