Abstract
Exocyclic DNA adducts are generated in cellular DNA by various industrial pollutants such as the carcinogen vinyl chloride and by endogenous products of lipid peroxidation. The etheno derivatives of purine and pyrimidine bases 3,N4-ethenocytosine (ɛC), 1,N6-ethenoadenine (ɛA), N2,3-ethenoguanine, and 1,N2-ethenoguanine cause mutations. The ɛA residues are excised by the human and the Escherichia coli 3-methyladenine-DNA glycosylases (ANPG and AlkA proteins, respectively), but the enzymes repairing ɛC residues have not yet been described. We have identified two homologous proteins present in human cells and E. coli that remove ɛC residues by a DNA glycosylase activity. The human enzyme is an activity of the mismatch-specific thymine-DNA glycosylase (hTDG). The bacterial enzyme is the double-stranded uracil-DNA glycosylase (dsUDG) that is the homologue of the hTDG. In addition to uracil and ɛC-DNA glycosylase activity, the dsUDG protein repairs thymine in a G/T mismatch. The fact that ɛC is recognized and efficiently excised by the E. coli dsUDG and hTDG proteins in vitro suggests that these enzymes may be responsible for the repair of this mutagenic lesion in vivo and be important contributors to genetic stability.
Keywords: ethenoadducts/base excision DNA repair/lipid peroxidation
A number of chemical carcinogens induces the formation of cyclic adducts in DNA. These include industrial chemicals such as vinyl chloride (1) and the widespread environmental compound ethyl carbamate (2, 3). Ethyl carbamate is a carcinogen that is metabolized to vinyl carbamate, then oxidized by cytochrome P450 to the electrophyle vinyl carbamate epoxide that reacts with RNA and DNA bases to form etheno-bridged adducts (4–6). Similarly, two oxidized metabolites of vinyl chloride, 2-chloroacetaldehyde and 2-chloroethylene oxide, predominantly react with purine and pyrimidine residues in DNA and RNA producing 1,N6-ethenoadenine (ɛA), 3,N4-ethenocytosine (ɛC), N2,3-ethenoguanine (ɛG), and 1,N2-ethenoguanine (1,N2-ɛG) (7, 8). Furthermore, the generation of exocyclic DNA adducts by products of membrane lipids peroxidation has been demonstrated (9, 10). The level of ɛC present in human liver has been found to be 2.8 ± 0.9 per 107 bases (11).
The mutagenic potential of cyclic DNA lesions has been established. In Escherichia coli and simian kidney cells, ɛC mostly produces ɛC⋅G to A⋅T transversions and ɛC⋅G to T⋅A transitions (12, 13). For single-stranded shuttle vestor containing a single ɛC residue the targeted mutation frequency yields a 2% in E. coli, 32% in SOS-induced E. coli cells, and 81% in simian kidney cells (13). For comparison, the apparent mutation frequency measured in E. coli for a single C8-hydroxyguanine residue in double-stranded M13 phage DNA is 0.3% (14). In mammalian cells, ɛA residues mainly lead to ɛA⋅T to G⋅C transitions (15). In bacterial systems, ɛG has miscoding properties producing ɛG⋅C to A⋅T transitions (16), and 1,N2-ɛG leads to G⋅C to A⋅T transitions (17). The repair of ɛA and ɛG adducts by DNA glycosylases present in crude cells extracts has been described (18, 19). The excision of ɛA by pure 3-methyladenine-DNA glycosylases of various origins have shown that the human enzyme is by far the most efficient (20). The purified AlkA protein releases ɛG when present in DNA (21).
An enzymatic activity repairing ɛC has been identified and partially purified from human cells and shown to be different from the ANPG protein (22, 23) but has not been characterized at the molecular level. Surprisingly, until now there was a complete lack of information about the repair of ɛC in prokaryotes.
In the present study, using an ɛC-containing duplex oligonucleotide, we purified to homogeneity an ɛC-DNA glycosylase (ɛCDG) activity from E. coli cell extract. It was identified as the double-stranded uracil-DNA glycosylase (dsUDG) (24). The protein acts with an unusual efficacy. We have identified, by analogy (25, 26), the human mismatch-specific thymine DNA glycosylase (hTDG) (27) as the enzyme excising ɛC in human cells, also with a good efficiency. These observations suggest a possible role of these proteins in vivo to counteract the genotoxic effects of ɛC residues.
MATERIALS AND METHODS
Oligonucleotides.
The 34-mer oligonucleotide 5′-AAATACATCGTCACCTGGGXCATGTTGCAGATCC-3′, where at position 20, X = ɛC, U, or T, was purchased from Genset (Paris). This sequence previously was used to identify the ethenoadenine and the hypoxanthine-DNA glycosylases (20, 28). These sequences will be referred to as (ɛC-34), (U-34), or (T-34). The 34-mer oligonucleotide containing ɛC, U, or T was 32P-labeled at the 5′-end by T4 polynucleotide kinase or at the 3′-end by terminal transferase, yielding [32P] 5′-end- or 3′-end-labeled ɛC-34, U-34, or T-34 oligonucleotides. Four complementary oligonucleotides, containing dA, dG, dC, or T opposite to X were synthesized by E. Lescot (this laboratory). The duplex oligonucleotides, made by annealing (ɛC-34), (U-34), or (T-34) with the complementary oligonucleotides as already described (28), will be referred to as ɛC-34/G, ɛC-34/A, ɛC-34/T, ɛC-34/C or U-34/G, U-34/A, U-34/T, U-34/C or T-34/G, T-34/A, T-34/T, T-34/C when the base opposite to the adduct is G, A, T, C, respectively. We also used the following 34-mer oligonucleotide containing thymine at position 19 (T-19): 5′-CGGTATCCACCAGGTCATTAATAACGATGAAGCC-3′ annealed to a complementary oligonucleotide containing dG at position 19 (synthesized by E. Lescot of this laboratory). This duplex oligonucleotide will be referred to as T-19/G.
Enzymes.
Xth protein, terminal transferase, and molecular biology products were purchased from Boehringer Mannheim. T4 polynucleotide kinase was purchased from New England Biolabs. The purification of the E. coli FPG protein (29), Nfo protein (30), UNG, Nth protein (31), Tag, and AlkA protein (32, 33) was performed as described. The ANPG40 (34), ANPG60 (35), and APDG60 (32) proteins were purified to apparent homogeneity from extracts of E. coli BH290 (tag, alkA) harboring plasmids containing the ANPG40, ANPG60, and APDG60 cDNAs, respectively. The activity of the various proteins was tested by using their classical substrates and was checked just before their use.
DNA Glycosylase Assays.
The DNA glycosylase assay and analysis of the products of the reaction were performed as described (28) but using ɛC-34/G as substrate. Incubations were made at 30°C for the human enzyme and at 37°C for the bacterial enzyme, unless otherwise stated. The products released by the enzymes were characterized by HPLC using a Beckman System Gold equipped with a C18 μBondapak column. The column was isocraticaly eluted at 1 ml/min with 50 mM NH4H2PO4 (pH 4.5) containing 10% methanol (vol/vol). The elution of the products was monitored by UV absorption at 270 nM and 280 nM.
Enzymes Purification.
Purification of the bacterial ɛCDG. To purify the ɛCDG from bacterial cells, we chose the E. coli strain RZ1032 (ung, dut). Cell-free extract was prepared by using freezing-thawing cycles without lysozyme. After overnight culture (1 liter), the cells were harvested by centrifugation and washed with 250 ml of buffer A [0.3 M Tris⋅HCl, pH 8.0/5 mM EDTA/0.1 mM phenylmethylsulfonyl fluoride (PMSF)]. They were resuspended in the same buffer (1 g cells/8 ml buffer) and stored frozen at −20°C. Frozen cells were thawed in ice-water bath for 1 h and then heated for 10 min at 37°C. Then the mixture was placed in a dry ice-ethanol bath for 15 min. This procedure was repeated 3–4 times, and the lysate was clarified by centrifugation (10,000 g for 10 min at 4°C). The supernatant was filtered through 22 μm (fraction I) and made 1.7 M in ammonium sulfate. The resulting precipitate was removed by centrifugation, and the supernatant (fraction II) was applied on a Phenyl-Sepharose (Pharmacia) column (1.5 × 3.5 cm) and rinsed with buffer B (buffer A containing 1.7 M ammonium sulfate). The proteins were eluted by using a linear gradient (total volume 60 ml), 0–100% of buffer C (20 mM Tris⋅HCl, pH 8.0/2 mM EDTA/2.5 mM β-mercaptoethanol/0.1 mM PMSF/5% glycerol). Fractions containing ɛCDG activity were pooled and dialyzed against buffer D (as buffer C, but without glycerol). The dialyzed solution (fraction III) was applied on a Mono Q HR 5/5 FPLC column using an FPLC system (Pharmacia). The flow-through (fraction IV) was collected and loaded on a Mono S HR 5/5 column. The column was rinsed with buffer D, and a gradient from 0 to 800 mM NaCl in buffer D (15 ml, 30 min, 0.5 ml/min) was used to develop the column. Fractions containing the ɛCDG activity were supplemented with glycerol (50%) and stored at −20°C.
Purification of the hTDG protein.
The plasmid DNA pT7-hTDG containing the TDG cDNA coding for the hTDG (27) was provided by J. Jiriçny (Institute for Medical Radiobiology, Zurich, Switzerland). The purification procedure was similar to the method described by Neddermann et al. (27) but with modifications. A 2-liter culture of E. coli BL21 (DE3) containing pT7-hTDG was grown at 30°C to A600 = 0.9. It then was cooled to 22°C and induced with 1 mM isopropyl β-d-thiogalactoside for 12 h at 22°C. Bacterial cells were collected by centrifugation, washed with TEG buffer (25 mM Tris, pH 8.0/0.1 mM phenylmethylsulfonyl fluoride/2 mM EDTA/10% glycerol/2.5 M β-mercaptoethanol). They were resuspended in 80 ml of the same buffer and stored at −20°C. The cells were lysed by sonication at 4°C, and the lysate was adjusted to 0.1 M NaCl and centrifuged (30,000 rpm for 60 min, in a Beckman 42.1 rotor). The supernatant (80 ml, fraction I) was loaded on a column of DEAE-Sepharose Fast Flow Resin (Pharmacia) (1.44 × 4.6 cm). The G/T mismatch-specific DNA glycosylase activity was recovered in the flow-through fraction whereas most of E. coli ɛCDG activity remained on the QMA column. The flow-through fraction (fraction II) then was applied on a POROS 20 HS (1 ml column) (Boehringer Mannheim) equilibrated with TEG buffer containing 0.1 M NaCl. The human protein was eluted by using a linear gradient of 0.1 to 1 M NaCl in TEG buffer. The fractions containing G/T mismatch-specific DNA glycosylase activity were collected and diluted with TEG buffer to 0.1 M NaCl (fraction III) and loaded on Mono Q HR 5/5 column (Pharmacia). Again, the G/T mismatch-specific DNA glycosylase activity did not stick to the column and was recovered in the flow-through fraction (fraction IV). Fraction IV was directly loaded on a Mono S HR 5/5 column (Pharmacia). The G/T mismatch-specific DNA glycosylase activity was eluted by using a 0.1–0.8 M NaCl linear gradient developed for 30 min at 0.5 ml/min. The hTDG activity peak eluted at about 0.5 M NaCl.
RESULTS
Identification of an Activity Excising ɛC in Crude Extracts of E. coli.
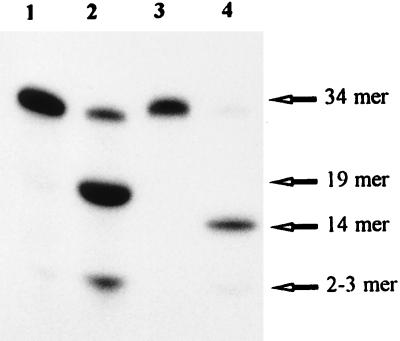
Although the ɛCDG activity has been identified in mammalian cells extracts, such enzyme has never been described in E. coli extracts. As shown in Fig. 1, incubation of the 5′-end-labeled oligonucleotide ɛC-34/G containing a single ɛC residue at position 20, with crude extracts from E. coli AB1157 and subsequent analysis by electrophoresis on a denaturing polyacrylamide gel, shows the appearance of reaction products migrating at the position of a 19-mer and a lower migrating band presumably generated by a nonspecific exonuclease activity present in E. coli extracts. When a 3′-end-labeled ɛC-34/G duplex oligonucleotide is used as substrate, the product of the reaction migrates at the position of a 14-mer. These results suggest that the incision occurs by the action of a DNA glycosylase followed by the cleavage of DNA by an endonuclease at abasic site. To identify the enzyme responsible for the repair of ɛC in E. coli, we investigated whether this lesion was a substrate for previously characterized DNA repair enzymes. The ɛCDG activity was checked in crude extracts of various E. coli strains deficient in the following DNA repair proteins: RZ1032 (ung), GC4803 (tag, alkA), BH20 (fpg), M182 (micA), BW372 (nth), BW528 (nfo, xth), BL101 (uvrA), NR9288 (mutD5), DH5α (recA), and JC7623 (recBC sbcB sbcC). All of the E. coli strains tested contained an activity incising the ɛC-34/G duplex oligonucleotide (data not shown), suggesting that the ɛCDG was an uncharacterized enzyme. Hence, the ɛC repair activity was further purified from E. coli cell extract, to identify the protein catalyzing this activity.
Figure 1.
Cleavage of oligonucleotides containing ɛC residues by E. coli cells extract. 5′-end or 3′-end 32P-labeled oligonucleotides containing ɛC residues were incubated with E. coli AB1157 cell extract. The products of the reaction were separated on a 20% denaturing PAGE. Lane 1, 5′-end 32P-labeled ɛC/G duplex oligonucleotide. Lane 2, as lane 1, but incubated with 5 μg of E. coli crude extracts for 10 min. Lane 3, 3′-end 32P-labeled ɛC/G duplex oligonucleotide. Lane 4, same as lane 3, but incubated with 5 μg of crude extracts for 10 min. The products of the reaction were analyzed by electrophoresis on a denaturing 20% polyacrylamide gel and visualized by using the PhosphorImager (model Storm 840). For details see Materials and Methods.
Purification of ɛC-Repair Activity from E. coli Cell Extract.
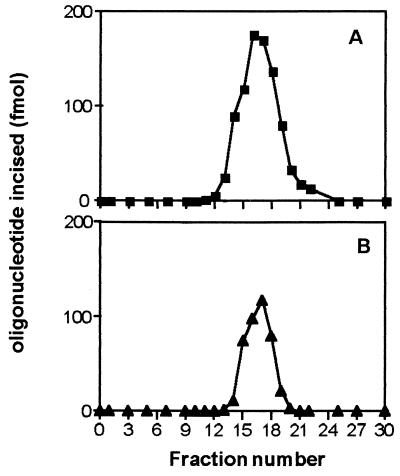
The ɛC repair activity excising ɛC from the ɛC-34/G was purified from E. coli strain RZ1032 (ung, dut), using the purification procedure described in Materials and Methods, including various columns that allowed us to separate the proteins according to their charge and hydrophobic properties. In the final step, the ɛC repair activity eluted from the Mono S column as a single symmetrical peak at a concentration of 0.43 M NaCl (Fig. 2A). Fractions containing the ɛC repair activity were analyzed by SDS/PAGE. The most active fraction was found to be purified to apparent homogeneity and consisted of a single polypeptide with an apparent size of about 22 ± 2 kDa (data not shown). This protein was used to obtain peptides for microsequencing.
Figure 2.
Distribution of ɛC and uracil-DNA glycosylase activities after FPLC chromatography on Mono S FPLC HR 5/5 column. The proteins containing the ɛCDG activity (fraction IV) were loaded on a Mono S FPLC HR 5/5 column and eluted by a linear NaCl gradient. (A) 250 fmol of the ɛC-34/G (■) duplex oligonucleotides were incubated with 1 μl from each column fraction for 10 min in a 50-μl reaction mixture. (B) 250 fmol of the U-34/G (▴) duplex oligonucleotides were incubated with 1 μl from each column fraction for 30 min in a 50-μl reaction mixture. The products of the reaction was analyzed as described in Fig. 1 and quantified by using the PhosphorImager (model Storm 840). For details see Materials and Methods.
ɛCDG Is an Enzymatic Activity of the E. coli dsUDG.
The partial trypsin proteolysis of ɛCDG yielded several peptides that were separated by HPLC. Two peptides were subjected to microsequencing. Their N-terminal residues were QLKPQEAHLLDYR and VIYQAGFTDR. Data bank searches of E. coli ORFs revealed that those peptide sequences correspond to the amino acid sequence of a 168-aa protein with a molecular weight of 18,673 Da (ref. 36 and G. Plunkett 3rd, deposited in GenBank in 1995, ECU28379, accession no. U28379) that is the dsUDG (24). This enzyme excises uracil residues from duplex DNA containing U/G mispair. Therefore, we have tested whether the ɛCDG and the dsUDG activities cochromatographed during the purification procedure. As shown in Fig. 2B, for the last step of the purification procedure, it is indeed the case.
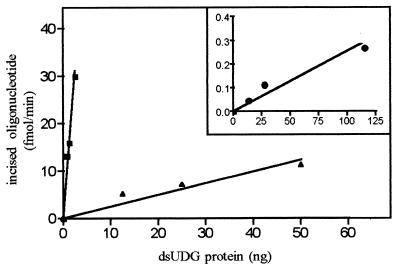
The kinetics of excision of ɛC and uracil residues present in a double-stranded oligonucleotide were compared. The results presented in Fig. 3 show that ɛC residues are excised 54-fold faster than uracil residues in the same experimental conditions. It should be recalled that the preparation of E. coli ɛCDG contains only the dsUDG activity, because it was purified from an E. coli ung− strain. From these results, we conclude that ɛCDG and dsUDG are two different enzymatic activities of the 168-aa protein, coded for in E. coli by the genetic locus ECU28379 (G. Plunkett 3rd, deposited in GenBank in 1995, accession no. U28379), and that the ɛCDG is a catalytic activity of the dsUDG protein that was yet unidentified.
Figure 3.
Activity of the E. coli dsUDG protein using as substrate ɛC-34/G (■), U-34/G (▴,) or T-19/G (•, Inset) as substrate. 5′-32P-labeled duplex oligonucleotide containing ɛC-34/G (■), U-34/G (▴), or T-19/G (•) mismatches (1 pmol in 100-μl reaction mixture) was incubated with increasing amounts of pure dsUDG protein for different periods of time in the presence of the Fpg protein. The products of the reaction were analyzed and quantified as described in Fig. 2. Each point represents the initial velocity of the enzymatic reaction. For details see Materials and Methods. Note the difference of scales in the Inset.
hTDG Excises ɛC When Present in DNA.
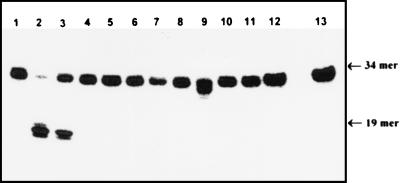
Because there is a high extent of homology between amino acid sequences of the dsUDG and the hTDG, we investigated whether the ɛCDG detected in human cell extracts (22) could be attributed to hTDG. Hence, the hTDG was purified from E. coli overexpressing the cloned hTDG by using as substrate an oligonucleotide containing the G/T mismatch. Because the substrate specificity’s of the hTDG and its bacterial homologue, the dsUDG protein, overlap, we have verified that the hTDG preparation was not contaminated by the dsUDG protein. The hTDG and dsUDG proteins can be completely separated during the purification procedure. During the first step of purification (the column of DEAE-Sepharose Fast Flow Resin), under the conditions used, the dsUDG protein sticks to the resin, whereas the hTDG protein is recovered in the flow-through fraction. Moreover we have established that during the next step (POROS 20 HS column, a cation exchange resin), any remaining dsUDG activity would have been efficiently separated from the hTDG activity by the linear gradient of NaCl, the dsUDG and the hTDG eluting at 0.35 and 0.50 M, respectively (data not shown). The human enzyme preparation could efficiently excise ɛC residues from the ɛC-34/G duplex oligonucleotide, when this residue is situated opposite to G (compare lanes 1 and 3 in Fig. 4). Because there are several examples showing that a modified base is recognized by more than one repair protein (37, 38) and that the efficacy of the enzymatic activity of a protein for a given substrate depends on its origin (20), we have investigated 11 pure repair proteins from E. coli and mammalian origin, involved in base excision repair, as potential candidates to repair ɛC residues. As shown in Fig. 4, only incubation of the ɛC-34/G with either the dsUDG or the hTDG, followed by Fpg treatment, leads to the incision of the duplex ɛC-34/G at the position of the modified base. AlkA, ANPG40, Fpg, Nth, Nfo, Xth, UNG, APDG60, and Tag proteins, although used in massive amounts to detect even marginal activity, do not act on the ɛC-34/G duplex under our experimental conditions (Fig. 4).
Figure 4.
Action of various E. coli and human DNA repair proteins on the 34-mer duplex ɛC/G oligonucleotide. The 5′-32P-labeled ɛC-34/G was incubated with an excess of the various pure repair protein at 37°C for 30 min (unless otherwise stated). Except for the control ɛC-34/G oligonucleotide and this oligonucleotide treated with Nth, Nfo, or Xth protein, the reactions were made in the presence of Fpg protein (50 ng) to reveal any abasic site generated by DNA glycosylases devoid of β-lyase activity. Lane 1, control ɛC-34/G oligonucleotide. Lane 2, as lane 1, but treated by E. coli dsUDG protein (5 ng). Lane 3, hTDG (150 ng, 30°C). Lane 4, AlkA (400 ng). Lane 5, ANPG40 (1.3 μg). Lane 6, Fpg protein (1 μg). Lane, 7, Nth protein (100 ng). Lane 8, Nfo (1.2 μg). Lane 9, Xth (4 nM, 10 min, 23°C). Lane 10, UNG (85 ng). Lane 11, APDG60 protein (1 μg). Lane 12, Tag I (350 ng). Lane 13, control as 1. The products of the reaction were analyzed as described in Fig. 1. For details see Materials and Methods.
E. coli dsUDG and hTDG Proteins Act on DNA Containing ɛC Residues as DNA Glycosylases.
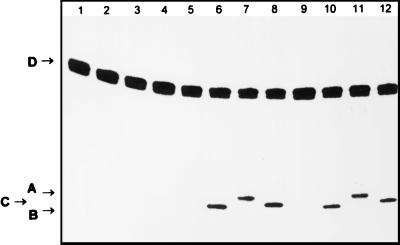
As shown in Fig. 5, treatment by enzymes nicking at the apurinic (AP) site (Fpg, Nth, or Nfo proteins) does not incise the ɛC-34/G duplex oligonucleotide (lanes 2–4), showing that there is no detectable AP site in the substrate. Upon treatment with dsUDG (lane 5) or hTDG (lane 9) proteins, no cleavage is detected, showing that our preparations are free of contaminant enzymes nicking at AP sites. However, treatment with enzymes that incise DNA at AP sites, after the action of dsUDG or hTDG proteins, generates a band migrating at the position of the 19-mer (lanes 6–8 and 10–12). Because the various enzymes used to nick at the AP site act through different catalytic mechanisms (39), the products of the enzymatic incision at the AP site of the duplex oligonucleotide migrate differently. The Fpg enzyme treatment cleaves the oligonucleotide at the AP site by a β-δ elimination mechanism generating a 32P-labeled fragment carrying a phosphate at the 3′-end (lanes 6 and 10). Nfo protein, which incises on the 5′-side of the AP site by a hydrolytic mechanism, generates a fragment that has a 3′OH termini and thus migrates slower than the fragment carrying a phosphate at the 3′-end (lanes 8 and 12) (40). The Nth protein incises at the abasic site by a β-elimination mechanism leaving on the 3′ end of the labeled 19-mer an α,β-unsaturated aldehyde that migrates slightly slower than the product of hydrolysis by Nfo (lanes 7 and 11) (40). These experiments show that the dsUDG and hTDG proteins generate AP sites in DNA containing ɛC residues and therefore act as DNA glycosylases.
Figure 5.
Mechanism of action of the dsUDG and hTDG proteins on ɛC/G oligonucleotide. The ɛC-34/G duplex oligonucleotide was incubated with dsUDG protein or hTDG protein, and subsequently treated or not with proteins nicking at AP sites to reveal abasic sites generated by dsUDG or hTDG proteins. The 5′-32P-labeled 34-mer ɛC-34/G duplex oligonucleotide is: lane 1, incubated at 37°C for 30 min; lanes 2, 6, and 10, incubated with 100 ng of Fpg protein at 37°C for 10 min; lanes 3, 7, and 11, incubated with 100 ng of Nth protein at 37°C for 10 min; lanes 4, 8, and 12, incubated with 100 ng of Nfo protein at 37°C for 10 min; lanes 5–8, incubated with 2 ng of dsUDG protein at 37°C for 10 min; lanes 9–12, incubated with 50 ng of hTDG protein at 30°C for 30 min. The products of the reaction were analyzed as described in Fig. 1. Arrow A indicates the 19-mer oligonucleotide containing an α,β-unsaturated aldehyde at the 3′-end. Arrow B indicates the 19-mer oligonucleotide containing a phosphate at the 3′-end. Arrow C indicates the 19-mer oligonucleotide containing 3′-OH termini. Arrow D indicates the 34-mer ɛC-34 oligonucleotide. For details see Materials and Methods.
In addition, the nature of the product excised from ɛC-4/G by the repair enzymes was characterized by analysis on HPLC as described in Materials and Methods. The product of excision eluted as a single peak at 11.9 min. Furthermore this product cochromatographed with an authentic sample of ethenocytosine. No material was detected at the position of the nucleoside (data not shown). These two independent experiments show that ɛC residues are excised by a DNA glycosylase mechanism.
The E. coli dsUDG Processes G/T Mismatch.
The ability of the hTDG and dsUDG to excise ɛC suggests that both proteins could have a very similar substrate specificity. Because hTDG recognizes substrates containing G/T, U/G, and ɛC/G mismatches, we investigated whether the dsUDG protein is endowed with a G/T mismatch-specific activity. To measure this activity, we used, as suggested by Sibghat-Ullah et al. (41), an oligonucleotide T-19/G where the T residue is in the context of TpG/T. In fact, (see Fig. 3 Inset) the dsUDG processes G/T mispairs in DNA, although much less efficiently than the U- and ɛC-containing substrates.
Comparison of the Kinetic Parameters for Excision of ɛC, Uracil, and Thymine from Duplex Oligonucleotides Containing ɛC/G, U/G, and G/T Mispairs, Respectively, by the dsUDG and the hTDG.
To evaluate the relative substrate specificity of the dsUDG for its various substrates, we measured the kinetic parameters for the excision of ɛC, uracil, and thymine residues, opposite to G in a double-stranded oligonucleotide. The results show that the best substrate for the E. coli dsUDG enzyme is, by far, ɛC as compared with uracil, because the specificity constant is 50 times lower for the latter (Table 1).
Table 1.
Kinetic constants of the E. coli dsUDG and hTDG proteins for the excision of ɛC, uracil, and thymine (opposite to guanine)
| Enzyme | Substrate* | Km, nM | kcat, min−1 | kcat/Km, min × nM−1 |
|---|---|---|---|---|
| dsUDG | ɛC/G† | 2.5 ± 1.6 | 0.95 ± 0.22 | 0.38 |
| U/G | 22.7 ± 8.6 | 0.17 ± 0.03 | 0.77 × 10−2 | |
| T/G | 26 ± 10 | (0.43 ± 0.06) ×10−3 | 0.17 × 10−4 | |
| hTDG | ɛC/G | 24.3 ± 14.9 | (9.2 ± 1.7) ×10−3 | 0.38 × 10−3 |
| U/G | 12 ± 5 | (21 ± 3) ×10−3 | 1.73 × 10−3 | |
| T/G | 12.8 ± 4.9 | (0.9 ± 0.1) ×10−3 | 0.071 × 10−3 |
Substrate concentration range 2.5–100 nM.
Substrate concentration range 0.1–6 nM.
In addition, Table 1 presents the kinetic constants measured for the hTDG acting on the same set of oligonucleotides. The comparison of the specificity constants for the enzyme using as substrate ɛC/G or U/G mismatches leads to the conclusion that for the human enzyme uracil and ɛC are equally good substrates and the most preferred substrates as compared with T/G mismatch.
Base Pair Specificity of the Ethenocytosine-DNA Glycosylases.
The dsUDG and the hTDG recognized ɛC only when present in a double-stranded oligonucleotide. No detectable excision of ɛC was observed when the lesion was present in a single-stranded oligonucleotide (data not shown). The base-pair specificity of both enzymes was measured by using duplex oligonucleotides containing mismatches generated by each of the four different bases opposite ɛC or uracil. In each case, the initial velocity of the excision of ɛC or uracil was measured. The results are presented in Table 2.
Table 2.
The influence of the base opposite to ɛC and/or uracil residues on the rates of excision by E. coli dsUDG and hTDG proteins
| Substrate | DNA glycosylase activity, %
|
|
|---|---|---|
| E. coli dsUDG | hTDG | |
| ɛC/G | 100 | 100 |
| ɛC/A | 46 | 26.5 |
| ɛC/T | 93 | 21 |
| ɛC/C | 61 | 71 |
| U/G | 100 | 100 |
| U/A | 1 | <0 |
| U/T | 84 | <0 |
| U/C | 94 | 2 |
The ɛC-34 mer and/or U-34 mer were annealed with the complementary 34-mer oligonucleotides to generate the following mismatches: ɛC/G, ɛC/A, ɛC/T, and ɛC/C and/or U/G, U/A, U/T, U/C. Enzymatic activity was measured as a function of time, and the initial velocities were determined. For details see Materials and Methods.
The excision of ɛC by the dsUDG or by hTDG does not show any strict preference, although the ɛC/G mismatch was the best substrate. At variance, the repair of uracil residues exhibits a marked preference according to the opposite base. In the case of the E. coli protein, uracil is excised from U/G, U/T, and U/C mismatches with a similar efficiency, whereas its excision from U/A is negligible. The hTDG protein strongly preferred U/G as a substrate, the repair of uracil in U/C is very negligible, and it is not excised from U/T and U/A. The striking fact is that ɛC residues are excised from all four mismatches with a comparable efficiency by both proteins.
DISCUSSION
Ethenobases have attracted much attention as critical candidates in the etiology of human cancers, because these adducts lead to misincorporation upon replication or transcription. Ethenoadducts in DNA could be formed either by exogenous sources (vinyl chloride, ethyl carbamate, etc.) or by products of lipid peroxidation such as trans-4-hydroxy-2-nonenal generated during the cellular metabolism (9, 10).
Because ethenobases are known to be promutagenic and genotoxic (12, 13, 15–17), they have to be removed from the genomic DNA. In E. coli and in Saccharomyces cerevisiae, the enzymatic activity excising ɛA has been identified as the 3-methyladenine-DNA glycosylase, the AlkA and Mag proteins, respectively (20), whereas proteins excising ɛC from these two organisms are unknown. Enzymatic activities excising ɛC and ɛA have been identified in mammalian crude cell extracts (18). The human protein binding to and excising ɛA has been partially purified (19) and identified as the ANPG protein (19, 20). Using partially purified proteins from HeLa cells and ANPG knockout mice, it has been shown that the enzymes excising ɛA and ɛC are two different proteins (22, 23).
In the present study, an ɛCDG has been purified to homogeneity from E. coli cells extract and was identified as the E. coli dsUDG (24), which is the homologue of the hTDG (27). However, the molecular weight of the purified ɛCDG/dsUDG, estimated by its mobility on PAGE under denaturing conditions (22 ± 2 kDa), differs from the calculated one based on the gene sequence (18.7 kDa). Moreover we have established that hTDG, the human homologue of dsUDG, also excises ɛC residues.
To compare the substrate specificity of the E. coli and the human enzymes, the kinetic parameters were measured for both enzymes, using three different duplex oligonucleotides containing different mismatches. The dsUDG protein acted on DNA substrates in the order ɛC/G ≫ U/G ≫ G/T. It should be emphasized that the most preferred substrate for dsUDG is, by far, ɛC. The enzyme has an extremely efficient kinetic constant acting on ɛC/G (kcat/Km = 0.4 min⋅nM−1), 52-fold higher than on U/G mismatch (kcat/Km= 0.77 × 10−2 min⋅nM−1), which is believed to be the physiological substrate of this enzyme. This suggests, if one extrapolates the in vitro results to the in vivo activity, that the main role of that glycosylase in vivo could be the repair of ɛC lesions in DNA. The physiological relevance of the U/G activity of the dsUDG has not yet been ascertained.
Moreover, the human protein hTDG, previously characterized as an enzyme repairing T in G/T mismatches, very efficiently repairs ɛC residues in DNA. The order of the hTDG preference for different DNA substrates is U/G > ɛC/G > T/G. The difference among the kinetics parameters measured for the excision of ɛC residues is less prominent for the human enzyme as compared with the dsUDG. This situation is different from what has been observed for the excision of ɛA, the mammalian enzymes (human and rat) being much more efficient than the prokaryotic ones (20). From the comparison of the kcat/Km values of the hTDG protein for each substrate, we conclude that uracil and ɛC are the most preferred substrates. The specificity of the human protein for uracil mismatch is 4.5 times higher than for ɛC, the kcat/Km value for thymine being much lower than for uracil and ɛC.
The lower kcat/Km value measured for the hTDG using an ɛC substrate, compared with the constant measured for the dsUDG, suggests that in vivo this enzyme may act as part of an efficient multiprotein complex. Ascertaining the role of hTDG in vivo will require further investigations using genetic approaches. The high promutagenic properties of ɛC in mammalian cells (13) and its repair by hTDG lead to the possibility of identifying a human genetic disease associated with a deficient hTDG activity.
The excision of T in a G/T mismatch by dsUDG adds a substrate to this enzyme. The extremely low constant measured (kcat/Km = 1.7 × 10−5) suggests that this enzymatic activity does not have a real biological significance. However, this substrate, in conjunction with the known structure of duplexes containing ɛC/T, ɛC/G, or ɛC/A mismatches (42–44), will be of interest for structural investigations aiming to elucidate the molecular mechanisms involved in the catalytic action of the dsUDG. Therefore, the human and the E. coli enzymes exhibit the same substrate specificity.
Repair of thymine and uracil residues by dsUDG and hTDG proteins strongly depends on the nature of the opposite base (see Table 2) (45). At variance, the repair of the ɛC residue is not markedly influenced by the opposite base. It should be noted that the efficiency of the excision of the ɛA residues by AlkA, MAG, and ANPG proteins does not depend on the nature of the opposite base (20). These facts raise the possibility that the processes involved in the recognition of the various modified bases could be different, the ɛC residue being recognized per se whereas the uracil and thymine residues are recognized through the structure of the mismatch. In fact, the U/A and the T/A are not repaired (Table 2 and ref. 45). One can speculate that the dsUDG and the hTDG directly recognize the ethenoadduct and flip out the ɛC residue into a specific pocket within their active site, similar to mechanism described for the UNG protein (46).
In conclusion, our results show that the DNA repair activity excising ɛC is associated in E. coli with the dsUDG and in human cells with the thymine-DNA glycosylase. The kinetics data demonstrate that ɛC is the most preferred substrate for both enzymes. The association of the mismatch-specific DNA glycosylase activity with the ɛCDG activity in the same protein is conserved during evolution, because it is observed in two unrelated species, human and E. coli. This result implies that a possible role of the human and bacterial enzymes in vivo could be to protect the integrity of the cellular genome from carcinogens and mutagens producing ɛC adducts in DNA.
Acknowledgments
We thank Dr. J. Jiriçny for the gift of the plasmid expressing the human pT7-hTDG gene and Dr. J. Derancourt (UPR 9008 Centre National de la Recherche Scientifique, Montpellier) for the determination of the amino acid sequence of the ɛCDG/dsUDG. We also thank Dr. O. Fedorova for invaluable help. This work was supported by grants from the European Commission (ENV4-CT97-0505), Comité Radioprotection-Electrecité de France, and Fondation Franco-Norvegienne. M.S. is the recipient of a grant from the Fondation pour la Recherche Médicale.
ABBREVIATIONS
- ɛC
3,N4-ethenocytosine
- ɛA
1,N6-ethenoadenine
- ɛG
N2,3-ethenoguanine
- 1
N2-ɛG, 1,N2-ethenoguanine
- ɛCDG
ɛC-DNA glycosylase
- dsUDG
double-stranded uracil-DNA glycosylase
- hTDG
human thymine-DNA glycosylase
- AP
apurinic
References
- 1.Green T, Hathway D E. Chem Biol Interact. 1978;22:211–224. doi: 10.1016/0009-2797(78)90126-6. [DOI] [PubMed] [Google Scholar]
- 2.Miller J A, Miller E C. Br J Cancer. 1983;48:1–15. doi: 10.1038/bjc.1983.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guengerich F P, Kim D-H. Chem Res Toxicol. 1991;4:413–421. doi: 10.1021/tx00022a003. [DOI] [PubMed] [Google Scholar]
- 4.Dahl G A, Miller J A, Miller E C. Cancer Res. 1978;38:3793–3804. [PubMed] [Google Scholar]
- 5.Leithäuser M T, Liem A, Steward B C, Miller E C, Miller J A. Carcinogenesis. 1990;11:463–473. doi: 10.1093/carcin/11.3.463. [DOI] [PubMed] [Google Scholar]
- 6.Park K K, Surh Y J, Steward B C, Miller J A. Biochem Biophys Res Commun. 1990;169:1094–1098. doi: 10.1016/0006-291x(90)92007-m. [DOI] [PubMed] [Google Scholar]
- 7.Bolt H M. Crit Rev Toxicol. 1988;18:299–309. doi: 10.3109/10408448809037469. [DOI] [PubMed] [Google Scholar]
- 8.Guengerich F P, Min K S, Persmark M, Kim M S, Humphreys W G, Cmarik J L. In: DNA Adducts: Identification and Significance. Hemminki K, Dipple A, Shuker D E G, Kadlubar F F, Segerbäck D, Bartsch H, editors. Lyon, France: International Agency for Research on Cancer; 1994. pp. 57–72. [Google Scholar]
- 9.Chung F-L, Chen H-J C, Nath R G. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 10.El-Ghissassi F, Barbin A, Guichard Y, Bartsch H. Chem Res Toxicol. 1995;8:278–283. doi: 10.1021/tx00044a013. [DOI] [PubMed] [Google Scholar]
- 11.Marnett L J, Burcham P C. Chem Res Toxicol. 1993;6:785–771. doi: 10.1021/tx00036a005. [DOI] [PubMed] [Google Scholar]
- 12.Basu A K, Wood M L, Niedernhofer L J, Ramos L A, Essigmann J M. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 13.Moriya M, Zhang W, Johnson F, Grollman A P. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood M L, Esteve A, Morningstar M L, Kuziemko G M, Essigmann J M. Nucleic Acids Res. 1992;20:6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandya G A, Moriya M. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 16.Cheng K C, Preston B D, Cahill D S, Dosanjh M K, Singer B, Loeb L A. Proc Natl Acad Sci USA. 1991;88:9974–9978. doi: 10.1073/pnas.88.22.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langouët S, Müller M, Guengerich F P. Biochemistry. 1997;36:6069–6079. doi: 10.1021/bi962526v. [DOI] [PubMed] [Google Scholar]
- 18.Oesch F, Adler S, Rettelbach R, Doerjer G. In: The Role of Cyclic and Nucleic Acid Adducts in Carcinogenesis and Mutagenesis. Singer B, Bartsch H, editors. New York: Oxford Univ. Press; 1986. pp. 373–379. [Google Scholar]
- 19.Singer B, Antoccia A, Basu A K, Dosanjh M K, Fraenkel-Conrat H, Gallagher P E, Kusmierek J T, Qiu Z H, Rydberg B. Proc Natl Acad Sci USA. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saparbaev M, Kleibl K, Laval J. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matijasevic Z, Sekiguchi M, Ludlum D B. Proc Natl Acad Sci USA. 1992;89:9331–9334. doi: 10.1073/pnas.89.19.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hang B, Chenna A, Rao S, Singer B. Carcinogenesis. 1996;17:155–157. doi: 10.1093/carcin/17.1.155. [DOI] [PubMed] [Google Scholar]
- 23.Hang B, Singer B, Margison G P, Elder R H. Proc Natl Acad Sci USA. 1997;94:12869–12874. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallinari P, Jiriçny J. Nature (London) 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 25.Seeberg E, Eide L, Bjoeras M. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 26.Slupska M M, Baikalov C, Luther W M, Chiang J-H, Wei Y-F, Miller J H. J Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan J J, Wiebauer K, Jiriçny J. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 28.Saparbaev M, Laval J. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boiteux S, O’Connor T R, Lederer F, Gouyette A, Laval J. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 30.Levin J D, Johnson A W, Demple B. J Biol Chem. 1988;263:8066–8071. [PubMed] [Google Scholar]
- 31.Graves R J, Felszenszwalb I, Laval J, O’Connor T R. J Biol Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 32.O’Connor T R, Laval F. EMBO J. 1990;9:3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tudek, B., VanZeeland, A. A., Kusmierek, J. T. & Laval, J. (1998) Mutation Res., in press. [DOI] [PubMed]
- 34.O’Connor T, Laval J. Biochem Biophys Res Commun. 1991;176:1170–1177. doi: 10.1016/0006-291x(91)90408-y. [DOI] [PubMed] [Google Scholar]
- 35.O’Connor T R. Nucleic Acids Res. 1994;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borodovsky M, McIninch J D, Koonin E V, Rudd K E, Medigue C, Danchin A. Nucleic Acids Res. 1995;23:3554–3562. doi: 10.1093/nar/23.17.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 38.Melamede R J, Hatahet Z, Kow Y W, Ide H, Wallace S S. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 39.Dodson M M, Michaels M L, Lloyd R S. J Biol Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 40.Bailly V, Verly W G, O’Connor T, Laval J. Biochem J. 1989;262:581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sibghat-Ullah, Gallinari P, Xu Y-Z, Goodman M F, Bloom L B, Jiriçny J, Day R S., III Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 42.Korobka A, Cullinan D, Cosman M, Grollman P A, Patel D J, Eisenberg M, de los Santos C. Biochemistry. 1996;35:13310–13318. doi: 10.1021/bi9605696. [DOI] [PubMed] [Google Scholar]
- 43.Cullinan D, Korobka A, Grollman P A, Patel D J, Eisenberg M, de los Santos C. Biochemistry. 1996;35:13319–13327. doi: 10.1021/bi9605705. [DOI] [PubMed] [Google Scholar]
- 44.Cullinan D, Johnson F, Grollman P A, Eisenberg M, de los Santos C. Biochemistry. 1997;36:11933–11943. doi: 10.1021/bi9705725. [DOI] [PubMed] [Google Scholar]
- 45.Neddermann P, Jiriçny J. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 46.Savva R, McAuley-Hecht K, Brown T, Pearl L. Nature (London) 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]