Abstract
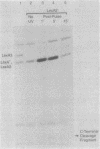
LexA2 repressor was partially inactivated after mitomycin C or UV light treatment in a recA+ or recA85(Prtc) (protease constitutive) host background. LexA2 protein was cleaved, but the reaction was slower than that observed for LexA+ repressor. lexA2 had a C-to-T transition at nucleotide 461 (Thr-154 to Ile).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R., Ptashne M. The lexA gene product represses its own promoter. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1932–1936. doi: 10.1073/pnas.77.4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Edmiston S. H., Pacelli L. Z., Mount D. W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Harper J. E. Identification of the lexA gene product of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6147–6151. doi: 10.1073/pnas.76.12.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W., Yanisch-Perron C. R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W. The SOS regulatory system: control of its state by the level of RecA protease. J Mol Biol. 1983 Jul 15;167(4):791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- Markham B. E., Little J. W., Mount D. W. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981 Aug 25;9(16):4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci U S A. 1977 Jan;74(1):300–304. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W., Low K. B., Edmiston S. J. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet lght-induced mutations. J Bacteriol. 1972 Nov;112(2):886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacelli L. Z., Edmiston S. H., Mount D. W. Isolation and characterization of amber mutations in the lexA gene of Escherichia coli K-12. J Bacteriol. 1979 Jan;137(1):568–573. doi: 10.1128/jb.137.1.568-573.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. R., Mount D. W. Differential repression of SOS genes by unstable lexA41 (tsl-1) protein causes a "split-phenotype" in Escherichia coli K-12. J Mol Biol. 1987 Jan 5;193(1):27–40. doi: 10.1016/0022-2836(87)90623-1. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]