Abstract
A plasmid expressing the cloned bacteriophage T4 gene 42 gave the same levels of complementation of gene 42 mutants in a polarity-suppressing rho mutant as in a rho+ host. A reading frame likely corresponding to gene 42 and putative promoter and terminator sequences were identified in the partial sequence of the cloned fragment.
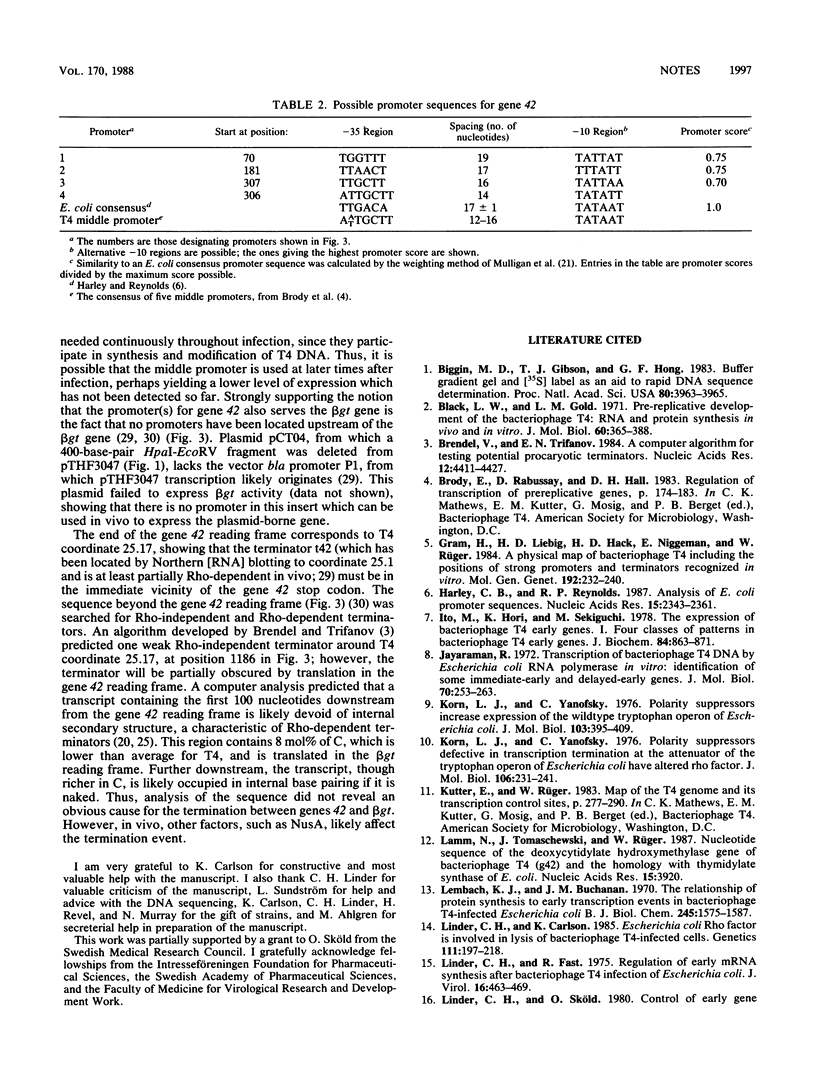
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black L. W., Gold L. M. Pre-replicative development of the bacteriophage T4: RNA and protein synthesis in vivo and in vitro. J Mol Biol. 1971 Sep 14;60(2):365–388. doi: 10.1016/0022-2836(71)90300-7. [DOI] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram H., Liebig H. D., Hack A., Niggemann E., Rüger W. A physical map of bacteriophage T4 including the positions of strong promoters and terminators recognized in vitro. Mol Gen Genet. 1984;194(1-2):232–240. doi: 10.1007/BF00383522. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Hori K., Sekiguchi M. The expression of bacteriophage T4 early genes. I. Four classes of patterns of expression in bacteriophage T4 early genes. J Biochem. 1978 Oct;84(4):865–871. doi: 10.1093/oxfordjournals.jbchem.a132198. [DOI] [PubMed] [Google Scholar]
- Jayaraman R. Transcription of bacteriophage T4 DNA by Escherichia coli RNA polymerase in vitro: identification of some immediate-early and delayed-early genes. J Mol Biol. 1972 Sep 28;70(2):253–263. doi: 10.1016/0022-2836(72)90537-2. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Yanofsky C. Polarity suppressors defective in transcription termination at the attenuator of the tryptophan operon of Escherichia coli have altered rho factor. J Mol Biol. 1976 Sep 15;106(2):231–241. doi: 10.1016/0022-2836(76)90082-6. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Yanofsky C. Polarity suppressors increase expression of the wild-type tryptophan operon of Escherichia coli. J Mol Biol. 1976 May 15;103(2):395–409. doi: 10.1016/0022-2836(76)90319-3. [DOI] [PubMed] [Google Scholar]
- Lamm N., Tomaschewski J., Rüger W. Nucleotide sequence of the deoxycytidylate hydroxymethylase gene of bacteriophage T4 (g42) and the homology of its gene product with thymidylate synthase of E. coli. Nucleic Acids Res. 1987 May 11;15(9):3920–3920. doi: 10.1093/nar/15.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembach K. J., Buchanan J. M. The relationship of protein synthesis to early transcriptive events in bacteriophage T4-infected Escherichia coli B. J Biol Chem. 1970 Apr 10;245(7):1575–1587. [PubMed] [Google Scholar]
- Linder C. H., Carlson K. Escherichia coli Rho factor is involved in lysis of bacteriophage T4-infected cells. Genetics. 1985 Oct;111(2):197–218. doi: 10.1093/genetics/111.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder C. H., Fast R. Regulation of early mRNA synthesis after bacteriophage T4 infection of Escherichia coli. J Virol. 1975 Sep;16(3):463–469. doi: 10.1128/jvi.16.3.463-469.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., Litchman B. L., von Hippel P. H. RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res. 1985 May 24;13(10):3739–3754. doi: 10.1093/nar/13.10.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T. W., Mathews C. K. T4 phage-coded deoxycytidylate hydroxymethylase: purification and studies on intermolecular interactions. Biochem Biophys Res Commun. 1977 Aug 8;77(3):898–904. doi: 10.1016/s0006-291x(77)80062-4. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M. Bacteriophage T4 gene expression. Evidence for two classes of prereplicative cistrons. J Biol Chem. 1973 Aug 10;248(15):5502–5511. [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M. Transcription and translation of prereplicative bacteriophage T4 genes in vitro. J Biol Chem. 1973 Aug 10;248(15):5512–5519. [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Revel H. R. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967 Apr;31(4):688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Thylén C. Controlling mechanisms for expression of the bacteriophage T4 beta-glucosyltransferase gene. J Gen Virol. 1987 Feb;68(Pt 2):253–262. doi: 10.1099/0022-1317-68-2-253. [DOI] [PubMed] [Google Scholar]
- Tomaschewski J., Gram H., Crabb J. W., Rüger W. T4-induced alpha- and beta-glucosyltransferase: cloning of the genes and a comparison of their products based on sequencing data. Nucleic Acids Res. 1985 Nov 11;13(21):7551–7568. doi: 10.1093/nar/13.21.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten J., Abelson J. The generation and analysis of clones containing bacteriophage T4 DNA fragments. J Mol Biol. 1980 Feb 25;137(2):235–248. doi: 10.1016/0022-2836(80)90327-7. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Tanyashin V. I., Murray N. E. Molecular cloning of fragments of bacteriophage T4 DNA. Mol Gen Genet. 1977 Nov 14;156(2):203–214. doi: 10.1007/BF00283493. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yeh Y. C., Greenberg G. R. Tetrahydrofolate-dependent labilization of the hydrogen atom on carbon 5 of 5'-deoxycytidylate, a step in the deoxycytidylate hydroxymethylase reaction. J Biol Chem. 1967 Mar 25;242(6):1307–1313. [PubMed] [Google Scholar]


