Abstract
Most human adenoviruses encode two virus-associated (VA) RNAs, VA RNAI and VA RNAII, that accumulate to high levels in the cytoplasm of infected cells. The function of VA RNAI in blocking the activation of the cellular kinase PKR is well known, but the role of VA RNAII is obscure. Herein we characterize and purify several human proteins that interact preferentially with VA RNAII in Northwestern blot assays. Two of these proteins were identified as RNA helicase A and NF90, a component of the heterodimeric nuclear factor of activated T cells (NFAT). They copurified with the smaller NFAT subunit, NF45, which did not bind VA RNAII, and with an unidentified protein, p97, which did bind VA RNAII. Both RNA helicase A and NF90 contain two copies of a double-stranded (ds) RNA binding motif and bind strongly to dsRNA. NF90 interacts with RNAs in the following order of affinity: dsRNA > VA RNAII > VA RNAI > single-stranded RNA. Furthermore, VA RNAII is more effective than VA RNAI as an inhibitor of RNA helicase activity. These data identify RNA helicase A and NF90 as cellular proteins with an affinity for dsRNA and other structured RNA molecules and suggest that their functions are subject to regulation by RNA ligands including VA RNAII.
Keywords: RNA helicase A/nuclear factor of activated T cells/NF90 subunit/NF45 subunit/protein kinase PKR
RNA molecules fulfill numerous structural, catalytic and regulatory roles in the cell. The virus associated (VA) RNAs are short (∼160 nucleotide), highly structured but single-stranded molecules that are actively synthesized by cellular RNA polymerase III. They accumulate to high concentrations in the cytoplasm of adenovirus-infected cells, predominantly during the late phase of virus infection (1). Most human adenovirus serotypes contain two VA RNA genes, encoding VA RNAI and VA RNAII, whereas the remainder carry a single gene of the VA RNAI type (2). VA RNAI counteracts a cellular antiviral defense mechanism that would otherwise lead to the shut-off of protein synthesis in infected cells. In particular, VA RNAI blocks the activation by double-stranded (ds) RNA of the interferon-inducible protein kinase PKR (previously known as DAI, P1, p68, or PK-ds) (3–6). Activated PKR inhibits protein synthesis by phosphorylating initiation factor eIF2 (7). Thus, VA RNAI maintains the functional integrity of the translation system in adenovirus-infected cells. It can also increase expression of proteins from transfected genes in uninfected cells (8, 9).
The function of VA RNAII is unclear. Its limited ability to block PKR activation (10, 11) argues against a role similar to that of VA RNAI, especially in light of its lesser accumulation in infected cells (12). Furthermore, a mutant virus that fails to produce VA RNAII grows as well as wild type in culture (13). These data are compatible with the view that VA RNAII does not play an important part in the adenovirus life cycle, but several lines of evidence favor the opposite point of view. (i) Although the VA RNAII− mutant virus is not defective in virus yield, the VA RNAI−/VA RNAII− double mutant grows about 10-fold less well than the VA RNAI− mutant (13–16). Moreover, virus production is delayed during the first 3 days of infection with the VA RNAII− mutant (13). (ii) VA RNAII is abundantly synthesized in infected cells (12), reaching about 107 molecules per cell (even though this is only about 10% of the level of VA RNAI). (iii) Although about 20% of human adenoviruses lack a VA RNAII gene, and some even seem to have lost it during recent evolution, the great majority of human serotypes (∼80%) possess a VA RNAII gene (2). (iv) Finally, and most compelling, the VA RNAII genes are at least as highly conserved as other VA RNA genes in sequence, and the secondary structures of their transcripts are also similar (2).
Such observations make it likely that the VA RNAII gene is biologically significant but shed little light on the process(es) in which it is involved. To address the possible biological function(s) of VA RNAII, we have investigated cellular proteins that interact with it. By using a Northwestern blotting assay, we detected eight proteins that bind preferentially to VA RNAII. Two of them were partially purified and identified by protein sequencing and antibody reactivity: one is RNA helicase A and the other is the large subunit (NF90) of the nuclear factor of activated T cells (NFAT or NF90/NF45). Their RNA binding properties were characterized, and VA RNAII was shown to inhibit RNA helicase A activity preferentially.
MATERIALS AND METHODS
Cells and Cell Extracts.
Human 293 cells (17), a line of adenovirus-transformed embryonic kidney cells, were grown in suspension culture. Cell extracts were prepared by Dounce homogenization and fractionated by standard procedures (18–20) to give nuclear extract (NE), 10,000 × g supernatant and pellet fractions (S10 and P10, respectively), and 100,000 × g supernatant and pellet fractions (S100 and P100, respectively), as diagrammed in Fig. 1A.
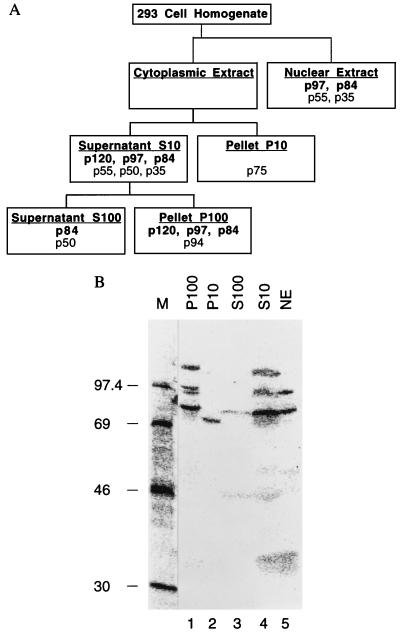
Figure 1.
Detection and subcellular localization of VA RNAII binding proteins. (A) Fractionation scheme showing the distribution of VA RNAII binding proteins. (B) Autoradiogram of a Northwestern blot, containing proteins from the indicated 293 cell fractions, probed with 32P-labeled VA RNAII. Lane M contains marker proteins whose molecular masses are indicated in kDa.
Northwestern Blotting and Immunoblotting.
Proteins were separated by SDS/PAGE (21) and then transferred to nitrocellulose membranes (Schleicher & Schuell BA85 membrane, 0.45-μm pore size) by electroblotting. For Northwestern blot analysis, membrane-bound proteins were subjected to denaturation and renaturation by sequential incubation with 6 M, 3 M, 1.5 M, 0.75 M, and 0.375 M guanidine hydrochloride in buffer C (20 mM Hepes⋅NaOH, pH 7.6/3 mM MgCl2/40 mM KCl/10 mM 2-mercaptoethanol). Membranes were then washed with buffer C, blocked with a 2% solution of Carnation nonfat dry milk in buffer C, and rinsed with buffer C containing 200 mM KCl. For probing with radiolabeled RNA, membranes were incubated for 1 h with 32P-labeled RNA (5 to 10 × 104 Cerenkov counts per min per ml) in buffer C containing 200 mM KCl, 0.1% Triton X-100, and 500 μg of heparin per ml. After washing with buffer C containing 200 mM KCl and 0.01% Triton X-100, the membranes were air-dried and exposed to film with enhancer screens for autoradiography or BAS 2000 Fuji Phosphoimager for imaging. For immunoblots, membranes were blocked with 5% Carnation nonfat dry milk in TBST (20 mM Tris⋅HCl, pH 7.6/150 mM NaCl/0.1% Tween 20) and then with 5% BSA in TBST. The membranes were probed with primary antibody (1:1,000 dilution) in 5% BSA/TBST for 1 h, rinsed with TBST, reblocked with 2% BSA/TBST, and then incubated with secondary antibody for 1 h. After washing with TBST, bound antibody was detected by enhanced chemiluminescence. Antibodies against human RNA helicase A were provided by Chee-Gun Lee (Sloan–Kettering Institute, New York) and against NF90 and NF45 by Peter Kao (Stanford University).
Purification of VA RNA Binding Proteins.
All procedures were carried out at 0–4°C with buffers containing 20% glycerol, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml, 1 μg of pepstatin A per ml, 0.2 mM phenylmethylsulfonyl fluoride, and 0.5 mM DTT. The S10 fraction of 293 cells was loaded onto a column of poly(I)⋅poly(C)-agarose resin (Pharmacia Biotech) equilibrated with buffer A (20 mM Hepes⋅NaOH, pH 7.6/3 mM MgCl2/200 mM KCl). The column was washed with buffer A containing 0.01% Triton X-100 and material was eluted with a linear gradient of 0.25–2 M NaCl in the same buffer.
Protein Sequence Analysis.
Peptide sequences were obtained from proteins separated by SDS/PAGE as described (22), except that the Tween concentration was 0.05%. Briefly, the proteins (3–10 pmol) were located by staining with Coomassie blue G (Sigma), excised, and digested in gel slices with Achromobacter protease I (Wako) for 20 h at 30°C. Peptides were separated on a C18 column and sequenced with an Applied Biosystems model 494 instrument. Initial yields averaged 3 pmol (range, 1.1–5.6 pmol).
Preparation of 32P-Labeled RNAs.
VA RNA probes were transcribed from DraI-linearized plasmids pT7VA2I and pT7VA2II (10) by using T7 RNA polymerase in the presence of [α-32P]UTP (ICN Radiochemicals) and purified by sequential electrophoresis through denaturing and native polyacrylamide gels (10, 23). dsRNA probe of 104 bp was prepared by transcription of pBSII KS+ (Stratagene), with T3 and T7 RNA polymerases (24). After annealing and removal of single-stranded (ss) RNA by digestion with RNases A and T1, the dsRNA was purified by electrophoresis through a native 10% polyacrylamide gel. dsRNA substrates for RNA helicase assays were prepared in the same way except that the T7 RNA polymerase transcription template was linearized with EcoRI instead of PvuII, and single-stranded ends were not trimmed by RNase digestion. ssRNA was prepared by transcription of EagI-linearized pBSII KS+ with T3 RNA polymerase and purification through two gels as described above.
Binding of RNAs to NF90.
NF90 protein was adsorbed from the S10 fraction of 293 cells onto protein A-Sepharose beads (Pharmacia) carrying rabbit polyclonal anti-NF90 antibodies (25). The NF90-containing Sepharose beads were washed six times with IP buffer (20 mM Hepes⋅KOH, pH 7.9/0.5 mM DTT/0.2 mM EDTA/50 mM KCl/20% glycerol/0.01% Nonidet P-40/100 units of aprotinin per ml/1 mM phenylmethylsulfonyl fluoride), equilibrated with buffer B (26), then incubated with RNA probe (5 × 104 cpm) in buffer B containing 100 μg of calf liver tRNA per ml and 1,000 units of RNASIN (Promega) per ml. for 5 min at 30°C and a further 25 min at 4°C. The beads were washed four times with buffer B containing 10 μg of calf liver tRNA per ml, and the binding of RNA to immobilized NF90 was monitored by Cerenkov counting.
Helicase Assay and Immunodepletion.
The RNA helicase assay was carried out as described (27) by using 2.5 to 5 × 104 cpm of 32P-labeled dsRNA substrate in each reaction. The source of enzyme was fraction 24 of the poly(I)⋅poly(C)-agarose column (dialyzed against buffer A containing 20% glycerol) or purified human RNA helicase A (kindly provided by Chee-Gun Lee). For immunodepletion, column fraction 24 was treated with protein A-Sepharose beads carrying antibody directed against human RNA helicase A (28) or an irrelevant rabbit preimmune serum as a control by using a slight modification of the procedure used for NF90 immobilization. The supernatants were assayed directly for RNA helicase or subjected to further immunodepletion and then assayed.
RESULTS
VA RNAII interacts with cellular proteins. The function of VA RNAI was revealed through the analysis of the protein synthesis deficiency associated with the VA RNAI-negative viral genotype (1, 5, 13, 29). In the absence of a clear phenotype resulting from the lack of VA RNAII, at least in cultured cells, we examined its interaction with cellular proteins as an alternative strategy to approach the role(s) of this RNA. Cytoplasmic and nuclear fractions were prepared from human 293 cells and the cytoplasmic extract was further fractionated by centrifugation to give S10, P10, S100, and P100 fractions (Fig. 1A). These fractions were subjected to analysis by the Northwestern blotting technique: proteins separated in an SDS/polyacrylamide gel were transferred to nitrocellulose membrane and probed with 32P-labeled VA RNAII.
About eight bands were detected in the various fractions (Fig. 1B). The most prominent and reproducible of these migrated with apparent molecular masses of 120, 97, and 84 kDa, corresponding to proteins designated p120, p97, and p84, respectively. The p120 band was detected in the cytosolic fractions P100 and S10 (lanes 1 and 4) but not in the P10, S100, or nuclear extract fractions (Fig. 1B, lanes 2, 3, and 5), implying that p120 may be ribosome-associated. The p97 and p84 bands were present in the same two cytosol fractions (Fig. 1B, lanes 1 and 4) and in the nuclear extract fraction (Fig. 1B, lane 5), suggesting that these proteins either shuttle between the nucleus and cytoplasm or leach out of nuclei during fractionation. A weak band comigrating with p84 was also seen in the S100 fraction (Fig. 1B, lane 3). Additional bands included p94, which was seen only in the P100 fraction (Fig. 1B, lane 1); p75, the only band detected in the P10 fraction (Fig. 1B, lane 2); p55 and p35, shared by the nuclear extract and S10 fractions (Fig. 1B, lanes 5 and 4); and p50, common to the S10 and S100 fractions (Fig. 1B, lanes 4 and 3). The distribution of these VA RNAII- binding proteins is depicted in Fig. 1A.
To address the specificity of these interactions, we probed with VA RNAI. The same bands were detected but the intensity of the signal was greatly attenuated, indicative of preferential binding to VA RNAII (data not shown). Similar proteins were also detected in extracts of human KB cells, although the p97 band was notably stronger than in 293 cell extracts (data not shown). The strength of the p50 signal, which probably corresponds to the La protein (30, 31) was variable with both probes, but the interactions with p120, p97, and p84 were reproducible and these proteins bound preferentially to VA RNAII rather than VA RNAI.
Identification of p120, p84, and a Copurifying Protein, p42.
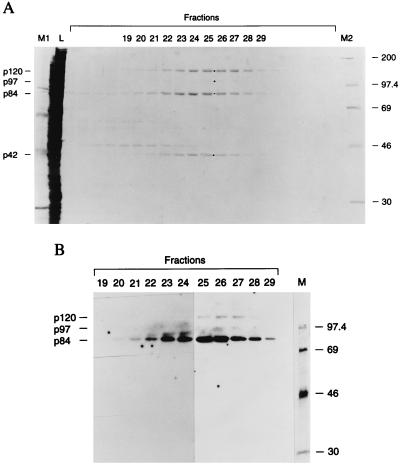
The 293 cell S10 fraction was chosen for protein purification because it contains the three prominent VA RNAII-binding proteins, p120, p97, and p84 and because the bulk of this RNA is cytoplasmic (12). These bands were all detected when Northwestern blots were probed with radiolabeled dsRNA (data not shown), suggesting that chromatography on poly(I)⋅poly(C)-agarose might be a useful purification step. Indeed, elution with a salt gradient gave a remarkably good one-step purification of the three proteins. As shown in the stained gel of Fig. 2A, proteins of the expected mobilities were present in fractions 20–29 (corresponding to about 0.6–1 M NaCl). The only other visible band, p42, coeluted with the peak of p120, p97, and p84. The latter three proteins were all detected when these fractions were analyzed for VA RNAII-binding activities by Northwestern blotting, but p42 was not detected (Fig. 2B).
Figure 2.
Purification of VA RNA binding proteins. (A) Copurification of p120, p84, and p42 through chromatography of the 293 cell S10 fraction over poly(I)⋅poly(C)-agarose. Eluted proteins were examined by SDS/gel electrophoresis and staining with Coomassie brilliant blue. Lane L contains a sample of the S10 fraction loaded on the column. (B) Detection of VA RNAII binding activities in column fractions by Northwestern blotting. Positions of the p120, p97, p84, and p42 bands are indicated with dots in A. Lanes M, M1, and M2 contain marker proteins with molecular masses in kDa.
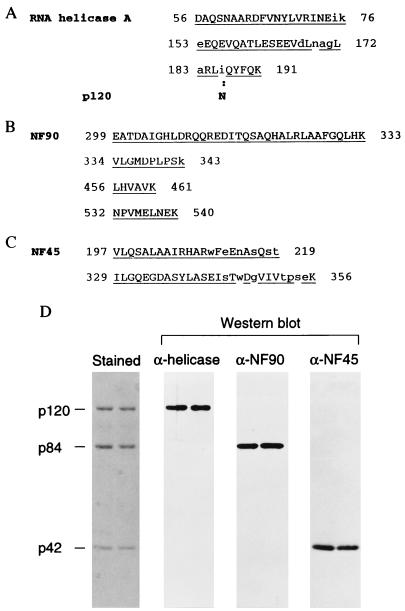
Proteins from a peak fraction were resolved by gel electrophoresis and subjected to peptide sequencing procedures. Data were obtained for p120, p84, and p42 and were compared with database information. Three peptide sequences from p120 matched human RNA helicase A (28) with a single discrepancy at residue 183, where asparagine was found instead of isoleucine (Fig. 3A). Asparagine is also reported at this position in the bovine homologue of human RNA helicase A (32). Four peptides from p84 matched the human protein NF90 (Fig. 3B), a component of the heterodimeric DNA-binding protein NF90/NF45 (33, 34). Two peptide sequences derived from p42 matched the NF45 sequence (Fig. 3C). Because p42 coeluted precisely with p84 from the poly(I)⋅poly(C) column but did not bind RNA in Northwestern analyses, we presume that p42 copurified through its interaction with p84.
Figure 3.
Protein identification. Amino acid sequences of peptides from p120 (A), p84 (B), and p42 (C) are matched with published sequences from human RNA helicase A, NF90, and NF45, respectively. The numbers indicate N-terminal and C-terminal amino acid positions for the published sequences. Identical residues are underlined; uppercase type denotes a clear sequencing signal; lowercase type represents amino acids that gave less distinctive signals or were not readable. A mismatch between p120 and RNA helicase A is indicated by boldface type. (D) Immunoblot analysis with antibodies directed against human RNA helicase A, NF90, or NF45. Proteins from a poly(I)⋅poly(C)-agarose fraction were resolved in an SDS/polyacrylamide gel and then examined by staining with Coomassie brilliant blue or by Western blot analysis as indicated.
To verify the identities of these proteins, they were resolved in an SDS/polyacrylamide gel, transferred to nitrocellulose membrane, and probed with antibodies to human RNA helicase A, NF90, and NF45. Comparison with the stained proteins showed that the antibodies specifically recognized p120, p84, and p42, respectively (Fig. 3D). Thus, these results identify the cellular VA RNA-binding proteins p120, p84, and the copurifying p42, with human RNA helicase A and the NF90–NF45 complex. Accordingly, the proteins will be referred to by their original names in the balance of this report.
Specificity of RNA Binding to Immobilized NF90 (p84).
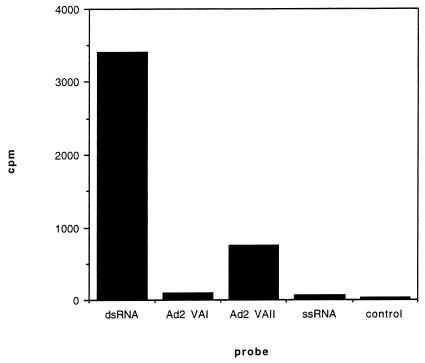
NF90, in a complex with NF45, binds to the NFAT DNA recognition element but has not previously been shown to bind RNA. To examine the RNA-binding activity of NF90, the protein was adsorbed from 293 cell S10 onto beads via anti-NF90 antibody. The beads were exposed to various 32P-labeled RNAs, unbound probe was removed, and bound RNA was monitored by assaying the retained radioactivity. Fig. 4 shows that NF90 efficiently bound dsRNA, whereas ssRNA gave a signal that was barely above background obtained in the absence of NF90. VA RNAII also bound efficiently, albeit to a lesser extent than dsRNA, whereas VA RNAI bound poorly. Similar results were obtained with chromatographically purified NF90 (data not shown). Thus, NF90 is able to bind dsRNA and highly structured ssRNAs such as VA RNAII; strikingly, in keeping with the results from Northwestern blotting, NF90 binds VA RNAII significantly better than VA RNAI even though both species have a high degree of secondary structure. The Northwestern blotting data make it likely that NF45 is not required for VA RNA binding to NF90.
Figure 4.
RNA binding to NF90 (p84). Sepharose beads carrying NF90 were exposed to 32P-labeled dsRNA, VA RNAI (Ad2 VAI), VA RNAII (Ad2 VAII), or ssRNA. The binding of RNA to immobilized NF90 was measured by the retention of radioactivity (cpm) on the beads. Data are averages of triplicate assays. Antibody was omitted for the control, which represents the average of single assays with each of the RNA probes.
Helicase Activity Is Inhibited by VA RNA.
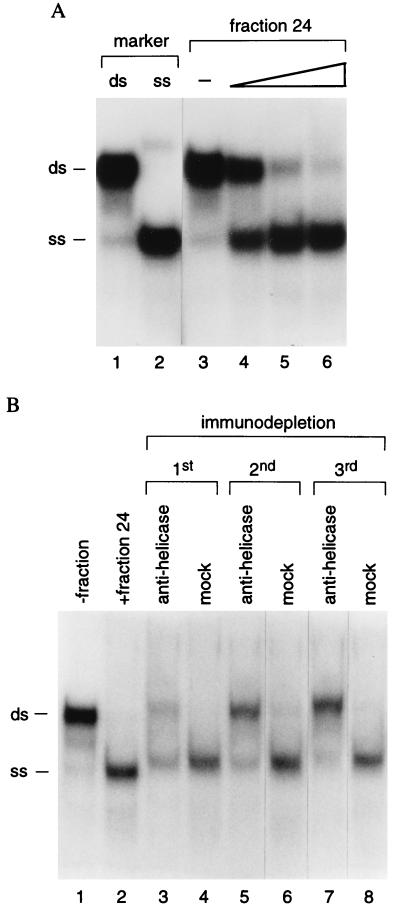
To determine whether the activity of RNA helicase A can be regulated by VA RNA, we first established that p120 purified by poly(I)⋅poly(C)-agarose chromatography possessed helicase activity. Fractions containing p120 were incubated with radiolabeled dsRNA and the reaction products were analyzed by gel electrophoresis to detect the conversion of dsRNA to ssRNA (Fig. 5A, lanes 1 and 2). Incubation with increasing volumes of fraction 24 led to a progressive decrease in the amount of dsRNA and concomitant appearance of ssRNA (Fig. 5A, lanes 3–6). Evidence that this is due to RNA helicase A was adduced from immunodepletion experiments (Fig. 5B). In the presence of fraction 24, dsRNA was converted to ssRNA (Fig. 5B, lanes 1 and 2). Serial immunodepletion with specific antibody progressively diminished the helicase activity of fraction 24 (Fig. 5B, lanes 3, 5, and 7), whereas control antibodies had no effect (Fig. 5B, lanes 4, 6, and 8), suggesting that RNA helicase A is the only RNA helicase active in the column fraction. As reported for human RNA helicase A (27), the RNA helicase activity of p120 is ATP-dependent: no unwinding of dsRNA occurred in the absence of ATP (Fig. 6A, lanes 4 and 5).
Figure 5.
RNA helicase activity is associated with p120. (A) 32P-labeled dsRNA substrate was incubated alone (lane 3) or with increasing amounts of fraction 24 from poly(I)⋅poly(C)-agarose chromatography (0.1, 0.3, and 0.5 μl for lanes 4–6, respectively). Lanes 1 and 2 contain untreated and boiled dsRNA, respectively. (B) RNA helicase assays were conducted in the absence (lane 1) or presence (lane 2) of fraction 24, with fraction 24 that had been depleted by treatment with antibody to human RNA helicase A (lanes 3, 5, and 7), or with mock-depleted (lanes 4, 6, and 8) fraction 24. Three sequential cycles of immunodepletion were conducted as indicated. The positions of dsRNA and ssRNA are marked.
Figure 6.
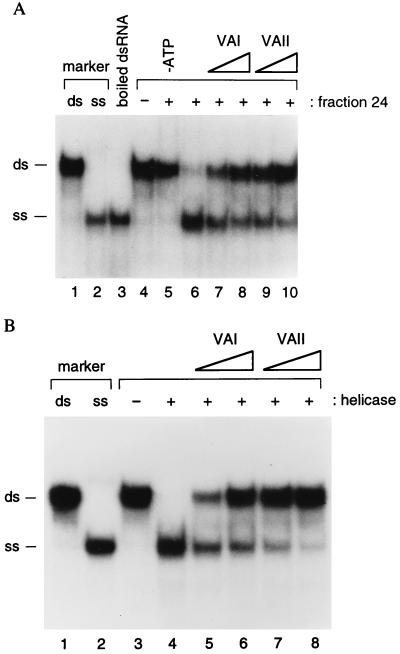
Inhibition of RNA helicase activity by VA RNAs. (A) Helicase assays with poly (I)⋅poly (C)-agarose fraction 24. Lane 6 contained a standard reaction. VA RNAI or VA RNAII were present in the reactions displayed in lanes 7–10 at 7.6 ng/ml (lanes 7 and 9) and 22.8 ng/ml (lanes 8 and 10). Control reactions lacking fraction 24 or ATP (lanes 4 and 5) were also conducted. Lane 3 contains boiled dsRNA substrate. (B) Helicase assays with purified human RNA helicase A. Lane 4 contained a standard reaction. VA RNAI or VA RNAII were present at 3.8 ng/ml (lanes 5 and 7) and 11.4 ng/ml (lanes 6 and 8). RNA helicase was omitted from the reaction of lane 3. Lanes 1 and 2 of A and B contain 32P-labeled dsRNA and ssRNA markers, and their positions are indicated.
Both VA RNAs reduced the unwinding of dsRNA (Fig. 6A, lanes 6–10). When tested at the same concentration, VA RNAII was approximately 3-fold more inhibitory toward helicase activity than VA RNAI. To verify that these results are not influenced by the presence of NF90/NF45 or other proteins in the poly(I)⋅poly(C)-agarose column fraction, the experiment was repeated with highly purified human RNA helicase A. Again, the VA RNAs reduced helicase activity and VA RNAII was approximately 3-fold more inhibitory than VA RNAI (Fig. 6B). It is important to note that the VA RNAs used in this study were purified through a procedure that is documented to remove dsRNA contaminants (23) and were free of dsRNA in PKR assays (data not shown). We conclude that VA RNA can inhibit the activity of RNA helicase A.
DISCUSSION
We have identified two cellular proteins, RNA helicase A and the NF90 component of NFAT, that can bind dsRNA and highly structured ssRNAs such as adenovirus VA RNAs. Both of these proteins are members of a growing family of proteins that contain the dsRNA-binding motif (dsRBM), and—like PKR—they both contain two tandem copies of the repeat (34, 35). This protein family includes PKR, Drosophila staufen, and Escherichia coli RNase III, for all of which convincing data exist demonstrating that the dsRBM is directly engaged in RNA binding (36–38), as well as other proteins that are known to interact with RNA (38). At present, it is an unverified but plausible assumption that the dsRBM is responsible for the observed interactions of RNA helicase A and NF90 with structured RNAs. If this proves to be the case, the results presented herein imply that the dsRBM does not bind structured RNAs indiscriminately. Both of these proteins appear to interact more strongly with VA RNAII than VA RNAI (Figs. 4 and 6), whereas PKR exhibits relatively stronger interactions with VA RNAI than VA RNAII (1, 10). Although the structures of two dsRBMs have been solved by NMR (38, 39), further analyses of the interactions of dsRBMs with RNAs will be required to understand the basis of the specificity.
Both RNA helicase A and NF90 participate in pivotal cellular control processes. RNA helicase A plays a central role in transcriptional regulation. It belongs to the DEAH subgroup of the DEAD box (DEAH/DexH) family of helicases and unwinds DNA as well as dsRNA (28, 40). Although RNA helicase A was first reported as a helicase that catalytically translocates in the 3′ to 5′ direction and did not unwind 5′-tailed DNA substrates significantly, our study shows that it can unwind 5′-tailed dsRNA substrates (Figs. 5 and 6). Its Drosophila homologue, the Maleless protein, plays an essential role in dosage compensation by up-regulating X chromosome transcription in the early stages of male fly development (28, 40). In human cells, it is associated with RNA polymerase II (pol II) and the cAMP responsive factor (CREB)-binding protein (CBP/p300) and functionally cooperates with this transcriptional coactivator to stimulate transcription (41). The NTPase/helicase activities of RNA helicase A and Maleless are essential for their transcriptional functions. Additional roles for RNA helicase A (for example, in replication or translation) have not been ruled out, and a posttranscriptional role in RNA export from the nucleus has been inferred (42). RNA helicase A is normally seen in the cell nucleus, but it appears to shuttle between the nucleus and cytoplasm in simian retrovirus-infected cells. This property may account for our detection of the helicase in the cytoplasmic fraction (Fig. 1).
Less is known about the functions of the NF90/NF45 complex. It was originally purified from T cells by virtue of its ability to bind to a DNA element, the ARRE, from the human interleukin 2 promoter (33, 34). Like other, unrelated, members of the NFAT family of transcriptional activators, its DNA binding activity was stimulated by ionomycin and phorbol 12-myristate 13-acetate, factors that stimulate T cell activation. That NF90 and NF45 play a role in transcription is suggested by immunodepletion experiments (33). Unlike other members of the NFAT family, however, the expression of NF90/NF45 is not limited to immune cells (34), raising the possibility that it participates in the transcription of a variety of genes. Despite their appearance in both the nuclear and cytoplasmic fractions (Fig. 1), NF90 and NF45 are predominantly detected in the nuclei by immunofluorescence. NF90 shares extensive homology with several proteins, including Xenopus dsRNA binding proteins (43), the mouse spermatid perinuclear RNA binding protein Spnr (44), and especially human M-phase phosphoprotein 4 (45). The latter is present throughout the cell cycle but is phosphorylated specifically during mitosis, suggestive of a cell-cycle-dependent function.
What is the significance of the interactions between these proteins and their RNA ligands? In the case of PKR, dsRNA serves as an activator of the latent kinase, whereas VA RNAI opposes the enzyme’s activation, thereby neutralizing one aspect of the cellular antiviral response. By analogy, VA RNAII may promote viral infection by regulating the properties of RNA helicase A and NF90. Support for this idea comes from the observations that VA RNAII binds selectively to NF90 and that it (and, to a lesser degree, VA RNAI) inhibits the dsRNA-unwinding action of RNA helicase A. Because these three proteins appear to interact with one another (I. Fierro-Monti, L. Parker, and M.B.M., unpublished results), it is tempting to speculate that they participate in a concerted regulatory mechanism that is influenced by one or both of the VA RNAs. Moreover, our data raise the possibility that other structured cellular and viral RNAs, such as the Alu and Ro RNAs and EBERs of Epstein–Barr virus, for example, may also exert control over cell functions through interactions with these RNA-binding proteins.
Acknowledgments
We thank Chee-Gun Lee, Jerard Hurwitz, and Peter Kao for materials and advice; our colleagues at Cold Spring Harbor Laboratory for help and encouragement; and Alain Verreault for discussions. This work was supported by Grants AI34552 and CA13106 from the National Institutes of Health.
ABBREVIATIONS
- VA
virus associated
- NFAT
nuclear factor of activated T cells
- ds
double-stranded
- ss
single-stranded
- dsRBM
dsRNA binding motif
References
- 1.Mathews M B, Shenk T. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Y, Mathews M B. J Virol. 1996;70:5083–5099. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Malley R P, Mariano T M, Siekierka J, Mathews M B. Cell. 1986;44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- 4.Kitajewski J, Schneider R J, Safer B, Munemitsu S M, Samuel C E, Thimmappaya B, Shenk T. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 5.Schneider R J, Safer B, Munemitsu S M, Samuel C E, Shenk T. Proc Natl Acad Sci USA. 1985;82:4321–4324. doi: 10.1073/pnas.82.13.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siekierka J, Mariano T M, Reichel P A, Mathews M B. Proc Natl Acad Sci USA. 1985;82:1959–1963. doi: 10.1073/pnas.82.7.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens M J. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 139–172. [Google Scholar]
- 8.Svensson C, Akusjärvi G. EMBO J. 1985;4:957–964. doi: 10.1002/j.1460-2075.1985.tb03724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufman R J, Murtha P. Mol Cell Biol. 1987;7:1568–1571. doi: 10.1128/mcb.7.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Mathews M B. J Virol. 1993;67:6605–6617. doi: 10.1128/jvi.67.11.6605-6617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori K, Juttermann R, Wienhues U, Kobayashi K, Yagi M, Sugimoto T, Tjia S T, Doerfler W, Hosokawa K. Virus Res. 1996;42:53–63. doi: 10.1016/0168-1702(95)01309-1. [DOI] [PubMed] [Google Scholar]
- 12.Söderlund H, Pettersson U, Vennström B, Philipson L, Mathews M B. Cell. 1976;7:585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- 13.Thimmappaya B, Weinberger C, Schneider R J, Shenk T. Cell. 1982;31:543–551. doi: 10.1016/0092-8674(82)90310-5. [DOI] [PubMed] [Google Scholar]
- 14.Bhat R A, Thimmappaya B. Nucleic Acids Res. 1984;12:7377–7388. doi: 10.1093/nar/12.19.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhat R A, Domer P H, Thimmappaya B. Mol Cell Biol. 1985;5:187–196. doi: 10.1128/mcb.5.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat R A, Thimmappaya B. J Virol. 1985;56:750–756. doi: 10.1128/jvi.56.3.750-756.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- 19.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dignam J D, Martin P L, Shastry B S, Roeder R G. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Wang R, Kobayashi R, Bishop J M. Proc Natl Acad Sci USA. 1996;93:8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellits K H, Pe’ery T, Manche L, Robertson H D, Mathews M B. Nucleic Acids Res. 1990;18:5401–5406. doi: 10.1093/nar/18.18.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manche L, Green S R, Schmedt C, Mathews M B. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao P N, Chen L, Brock G, Ng J, Kenny J, Smith A J, Corthesy B. J Biol Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 26.Clarke P A, Pe′ery T, Ma Y, Mathews M B. Nucleic Acids Res. 1994;22:4364–4374. doi: 10.1093/nar/22.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C G, Hurwitz J. J Biol Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 28.Lee C G, Hurwitz J. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 29.Reichel P A, Merrick W C, Siekierka J, Mathews M B. Nature (London) 1985;313:196–200. doi: 10.1038/313196a0. [DOI] [PubMed] [Google Scholar]
- 30.Lerner M R, Boyle J A, Hardin J A, Steitz J A. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- 31.Francoeur A M, Mathews M B. Proc Natl Acad Sci USA. 1982;79:6772–6776. doi: 10.1073/pnas.79.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Maacke H, Grosse F. J Biol Chem. 1995;270:16422–16427. doi: 10.1074/jbc.270.27.16422. [DOI] [PubMed] [Google Scholar]
- 33.Corthésy B, Kao P N. J Biol Chem. 1994;269:20682–20690. [PubMed] [Google Scholar]
- 34.Kao P N, Chen L, Brock G, Ng J, Kenny J, Smith A J, Corthésy B. J Biol Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 35.Gibson T J, Thompson J D. Nucleic Acids Res. 1994;22:2552–2556. doi: 10.1093/nar/22.13.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green S R, Mathews M B. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 37.St Johnston D, Brown N H, Gall J G, Jantsch M. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kharrat A, Macias M J, Gibson T J, Nilges M, Pastore A. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bycroft M, Grünert S, Murzin A G, Proctor M, St. Johnston D. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee K Y, Helbing C C, Choi K S, Johnston R N, Wang J H. J Biol Chem. 1997;272:5622–5626. doi: 10.1074/jbc.272.9.5622. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 42.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 43.Bass B L, Hurst S R, Singer J D. Curr Biol. 1994;4:301–316. doi: 10.1016/s0960-9822(00)00069-5. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher J M, Lee K, Edelhoff S, Braun R E. J Cell Biol. 1995;129:1023–1032. doi: 10.1083/jcb.129.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf J M. Mol Biol Cell. 1996;7:1455–1469. doi: 10.1091/mbc.7.9.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]