Abstract
Transposition of the maize Suppressor–mutator (Spm) transposon requires two element-encoded proteins, TnpA and TnpD. Although there are multiple TnpA binding sites near each element end, binding of TnpA to DNA is not cooperative, and the binding affinity is not markedly affected by the number of binding sites per DNA fragment. However, intermolecular complexes form cooperatively between DNA fragments with three or more TnpA binding sites. TnpD, itself not a sequence-specific DNA-binding protein, binds to TnpA and stabilizes the TnpA–DNA complex. The high redundancy of TnpA binding sites at both element ends and the protein–protein interactions between DNA-bound TnpA complexes and between these and TnpD imply a concerted transition of the element from a linear to a protein crosslinked transposition complex within a very narrow protein concentration range.
Keywords: maize/TnpA/TnpD/DNA-protein interaction
The maize Suppressor–mutator (Spm) element is a small, genetically complex transposon (1, 2). It encodes a single transcript that gives rise to several mature transcripts by alternative splicing (3). Two Spm-encoded proteins, TnpA (68 kDa) and TnpD (131 kDa), participate in transposition (1–4). TnpA binds to the subterminal repetitive regions of the Spm element, which contain multiple copies of sequence variants of the consensus TnpA binding site CCGACACTCTTA (5–7). Spm elements sustain both cis- and trans-acting mutations that affect the pattern of somatic mutation, delaying its timing in development and decreasing its frequency (8–11). The implication is that changes either in the element’s sequence or in the amount or structure of element-encoded proteins can affect the developmental timing and frequency of transposition.
The main impediment to studying the interaction of TnpA and TnpD with each other and with Spm DNA has been the difficulty of obtaining significant amounts of soluble intact proteins (4). Early binding studies carried out with the oligonucleotides and in vitro-translated TnpA led to the identification of a DNA-binding and a dimerization domain (4, 12). But, although it has been postulated that TnpA is responsible for the association of transposon ends, there is no supporting experimental evidence (1, 2). We have used soluble TnpA from Escherichia coli and extracts of plant cells expressing TnpD to analyze binding of the proteins to transposon sequences and interactions between the protein–DNA complexes. We also have studied protein interactions in a plant two-hybrid system (13). We show that, although the primary binding of TnpA to DNA is not cooperative, formation of intermolecular TnpA–DNA complexes exhibits cooperativity. We also show that TnpD interacts with TnpA and stabilizes its binding to DNA. These observations provide the basis for a biochemical explanation of mutations that change the developmental timing and frequency of transposition.
MATERIALS AND METHODS
TnpA Protein.
TnpA was over-expressed in E. coli as a fusion protein with the FLAG peptide at the N terminus (IBI FLAG expression system). To clone the TnpA cDNA in frame with the FLAG epitope, TnpA cDNA was transferred from pMS163 (14) into pBluescript II (KS+) (Stratagene) and then as a HindIII-XbaI fragment into the pFLAG-MAC plasmid to give pRR483, replacing 6 N-terminal amino acids with 14 amino acids, including the 8 amino acids of the FLAG peptide. TnpA expression in E. coli cells was induced with isopropyl β-d-thiogalactoside under conditions (0.1 mM isopropyl β-d-thiogalactoside, 28°C, 150 rpm, 20 hr) yielding >50% of the fusion protein in the soluble fraction. The protein was purified by antibody affinity chromatography as directed by the manufacturer. Purified protein was aliquoted and stored in 20 mM Tris (pH 7.6), 137 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and 20% glycerol at −70°C. Protein concentrations were determined by using the Bio-Rad protein assay kit as directed by the manufacturer. Purified TnpA had <5% contaminating E. coli proteins and was used directly for DNA binding studies.
DNA Fragments for Binding Studies.
The oligonucleotide TTAATTAAAGAATGTCGGGGCCGACACTCTTAATGGAAG containing two TnpA binding sites (boldface) in a tail-to-tail orientation corresponding to binding sites 2 and 3 at the 5′ end of Spm (Fig. 1a) was inserted into the EcoRV site of pBluescript II (KS+). TnpA binding sites were released as a 279-bp fragment with XhoI and PvuI (Fig. 1b). Deletion derivatives of the Spm promoter have been described (15). Digestion of the respective plasmids with SalI released fragments of 306, 241, 190, and 145 bp containing 9, 6, 3, and 1 binding site(s), respectively.
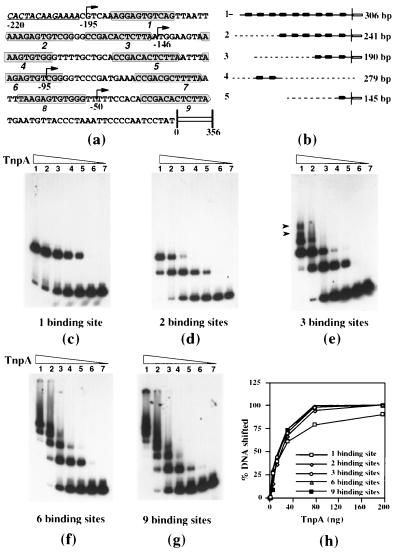
Figure 1.
Binding of TnpA to DNA fragments containing TnpA binding sites. (a) The 5′-terminal nucleotide sequence of Spm. Shaded boxes enclose the 12-bp TnpA binding motifs, whose orientation is indicated by the pointed end of the box. The underlined, italicized sequence is the terminal inverted repeat, and the open box represents the element’s GC-rich sequence (15). Arrows mark the deletion end points and the numbers indicate the distance (bp) of each from the transcription start site. (b) DNA fragments used in the mobility shift and ligation kinetics assays. Solid lines, Spm sequence; open box, the GC-rich sequence; broken lines, vector sequence; filled arrows, TnpA-binding sites; open arrow, terminal inverted repeat; vertical line, transcription start site. (c–g) DNA–protein complexes formed with different deletion derivatives of Spm promoter. A 279-bp fragment (b, lane 4) was used in d. All DNA fragments were labeled at both of the ends except those in d, which had only one labeled end. 32P-labeled DNA fragments were incubated with increasing amounts of TnpA in DNA-binding buffer, were fractionated on a 4% native acrylamide gel, and were detected by autoradiography. The arrows in e indicates the position of the additional complexes formed with target DNA containing three binding sites. (h) The percentage of shifted DNA in c–g is plotted as a function of the TnpA amount.
Assays of DNA Binding.
DNA was labeled by filling in restriction fragments by using [α-32P]dATP and Klenow fragment. The binding reactions were performed in 50 mM Tris (pH 7.8), 150 mM NaCl, 1 mM β-mercaptoethanol, 1 mM EDTA, and 10% glycerol in the presence of 1 μg of poly(dI-dC) and 5 μg of BSA in a total volume of 20 μl at 25°C for 20 min. Typically, 2 ng of labeled DNA was used with the amount of TnpA indicated in each figure. When tobacco cell or nuclear extracts were used for binding, 10 μg of protein was added, and the reaction was carried out in the presence of 2 μg of poly(dI-dC) and 2 μg of BSA. Where indicated, a 100-fold molar excess of unlabeled target DNA or the polylinker of bluescript KS+ were used as specific and nonspecific competitors in binding reactions. DNA–protein complexes were fractionated on a 4% polyacrylamide gel in Tris-glycine buffer at 30–35 mA for 4 hr at 4°C (16).
Ligation Kinetics Assay.
Intermolecular ligation was done as described (17). DNA fragments were isolated and labeled as described above. The binding reactions were performed in 20 μl containing 20 ng of labeled DNA, 120 ng of TnpA, 50 mM Tris⋅HCl (pH 7.4), 50 mM NaCl, 50 mM KCl, 0.02 mM EDTA, 30 μg/ml BSA, 2 mM ATP, 5 mM DTT, and 4% polyvinyl alcohol at room temperature for 20 min. A 3-μl aliquot was removed as the control and 1 μl of 100 mM MgCl2 and 2.5 units of T4 DNA ligase were added at room temperature. After 1, 2, 4, and 8 min, 3-μl aliquots were transferred to fresh tubes containing 3 μl of 2× stop solution (100 mM EDTA/1% SDS/0.02% bromophenol blue). DNA fragments were fractionated on a 1% agarose gel after incubation at 65°C for 15 min.
Tobacco Cell and Nuclear Extracts.
Cell and nuclear extracts were prepared from Nicotiana tabacum NT1 suspension cells (18). Cells expressing TnpD were established in suspension culture from transgenic N. tabacum SR1 containing a TnpD cDNA expressed from a cauliflower mosaic virus (CaMV) 35S promoter as described (14). The cell and nuclear extracts were prepared and stored essentially as described for Arabidopsis plants, except that 20 and 200 g wet weight of cells were used for cell and nuclear extracts, respectively (19).
DNase I Footprinting.
Binding reactions were performed as described above and were subjected to DNase I footprint analysis (16). After 20 min in binding buffer, 180 μl of 50 mM Tris (pH 7.8), 150 mM NaCl, 2 mM DTT, 2 μg/ml poly(dI-dC), and 50 μg/ml BSA were added, and incubation was continued for another 15 min. To this mixture, 5 μl of DNase I buffer (50 mM Tris, pH 7.8/150 mM NaCl/2 mM DTT/200 mM MgCl2/40 mM CaCl2) containing 20–50 units of DNase I (Boehringer Mannheim) was added, and the incubation was continued for 2 min at 25°C. The reaction was terminated by addition of 700 μl of DNase I stop solution (645 μl of 100% ethanol/5 μg of tRNA/50 μl of saturated ammonium acetate) and chilled in a dry ice/ethanol bath. The DNA was collected by centrifugation, was washed with 70% ethanol, was dried and dissolved in 10 μl of formamide loading buffer (0.05% wt/vol bromophenol blue/0.05% wt/vol xylene cyanol/20 mM EDTA in deionized formamide). The digestion products were analyzed on an 8% polyacrylamide 8 M urea gel.
Transient Expression Assays.
Diagrams of the DNA-binding domain plasmids (GAL4/TnpD), activation domain plasmids (TnpA/VP16), and the hybrid GAL4/CaMV promoter–luciferase (LUC) reporter plasmid are shown in Fig. 5. Plasmids pMS236 (GAL4), pMS245 [A(1–120)/VP16], pMS247 [A(1–422)/VP16], pMS249 [A(1–543)/VP16], and pDP1446 (GAL4/CaMV LUC reporter) and the chloramphenicol acetyl transferase reference plasmid pDC155 were described (13, 15). Plasmid pMS254 [GAL4/D(110–1134)] has amino acids 110-1134 of TnpD from CaMV 35S-tnpD (20) replacing VP16 (inserted as a BglII–XbaI fragment with a filled-in XbaI site) in pMS232 (13) cut with BglII and NcoI with a filled-in NcoI site to give plasmid pMS251. Plasmid pMS254 was made by blunt-end self-ligation of pMS251 cut and filled in at the BglII site. Plasmid pMS276 [GAL4/D(110–1122)] has amino acids 110-1122 of TnpD inserted as a ClaI–BbrPI fragment from pMS254 into pMS238 (13) cut with ClaI and NcoI (with filled-in NcoI site). This cloning caused a frameshift that replaced the 12 C-terminal amino acids of TnpD (RVAEEDSDADDF) by 4 amino acids (PWLT). The interaction of TnpA and TnpD was studied in tobacco NT1 suspension cells grown as described in Russell et al. (18). The assay has been described (13, 15).
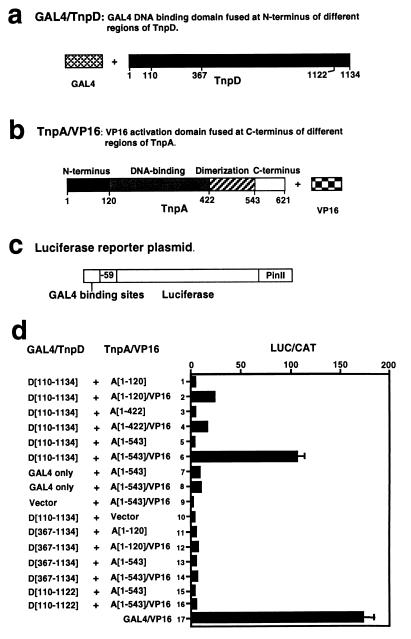
Figure 5.
Analysis of TnpA–TnpD interactions in a plant two-hybrid system. (a–b) Schematic representations of the components from which the fusion constructs were assembled. Each contained either the GAL4 DNA binding domain or the VP16 transcription activation domain but different segments of the TnpD and TnpA genes, respectively, as shown in d. All chimeric genes were expressed from the CaMV 35S promoter. (c) The structure of the LUC reporter gene, which contains five GAL4 DNA binding sites, a −59 to +2 minimal CaMV 35S core promoter, the LUC gene, and the potato proteinase inhibitor II terminator (PinII). (d) Relative expression of 1 μg GAL4–LUC reporter plasmid cobombarded into tobacco cells with 1 μg each of a plasmid containing the chimeric GAL4/TnpD gene and one containing a chimeric TnpA/VP16 gene (shown in a and b). Plasmids with the corresponding TnpA or GAL4 fragments were used as controls. The results are expressed as the ratio of LUC to chloramphenicol acetyl transferase activity and represent the average of three independent experiments, each comprising three replicates. D[110–1134], D[367–1134], and D[110–1122] designate the TnpD residues included in the GAL4 fusion protein; A[1–120], A[1–422], and A[1–543] designate the TnpA residues included in the VP16 fusion protein; vector designates a plasmid containing the CaMV 35S promoter, but no insert; GAL4/VP16 is a translational fusion of GAL4 binding domain and VP16 activation domain expressed from the CaMV 35S promoter.
RESULTS
Noncooperative Binding of TnpA.
We used soluble, purified TnpA expressed in E. coli and both natural and synthetic DNA fragments to study the binding of TnpA to DNA. Binding was assessed by the ability of TnpA to decrease the electrophoretic mobility of labeled oligonucleotides or DNA fragments with TnpA binding sites. Fig. 1a shows the complete promoter sequence with the TnpA binding sites and the endpoints of the deletions depicted in Fig. 1b. Fragment 4 is a synthetic oligonucleotide containing the second and third binding sites from the 5′ end of the element, previously reported to be the optimal TnpA binding sequence (12). Fragments 1, 2, 3, and 5 are deletion derivatives of the complete promoter (15). Broken lines represent plasmid sequences used to extend fragment length. As have others (12), we find that TnpA binds DNA fragments containing TnpA binding sites but not fragments lacking them (data not shown). Similarly, we find that bound TnpA can be displaced by the addition of excess DNA containing TnpA binding sites but not by the addition of DNA lacking them (12; data not shown).
Several inferences can be drawn from the results of the electrophoretic mobility shift experiments displayed in Fig. 1. First, it is likely that TnpA binds DNA as a monomer. DNA fragments containing one and two binding sites show one and two bands of retarded electrophoretic mobility, respectively, implying a 1:1 relationship between retarded species and the number of TnpA molecules bound. We see no evidence of additional bands that could represent TnpA dimers bound to a single binding site, as reported (12). Second, it is likely that each successive binding site is occupied independently. In Fig. 1d, the more rapidly migrating of the two retarded bands first increases and then decreases in intensity with increasing TnpA concentration. The same is true for DNA fragments containing larger numbers of binding sites, suggesting that TnpA molecules bind independently. The number of discrete DNA–protein complexes continues to increase with the number of binding sites per DNA fragment although individual bands become difficult to resolve as the number of binding sites increases beyond three (Fig. 1 f and g). When the fraction of the total shifted DNA is plotted against protein concentration (Fig. 1h), the curves are hyperbolic. Moreover, they are virtually superimposable for fragments containing different numbers of binding sites with the exception that less of the fragment containing a single binding site is shifted at saturation than of fragments with more binding sites, suggesting that this complex is less stable than the others (Fig. 1h). We conclude, therefore, in contrast to what has been reported (12), that the primary binding of TnpA to DNA is not cooperative.
Cooperative Formation of Intermolecular TnpA–DNA Complexes.
Complexes of progressively larger size are observed as the protein:DNA ratio increases. For small numbers of binding sites, the number of bands with reduced electrophoretic mobility observed in the presence of TnpA is the same as the number of binding sites, and the bands can be resolved easily for fragments with one, two, and three binding sites. Additional complexes of even lower mobility can be seen by using fragments with three or more binding sites (arrows in Fig. 1e). The complexes become larger and more prevalent as the number of binding sites increases (Fig. 1 f and g). Moreover, the fraction of the retarded DNA in large complexes increases with numbers of binding sites per DNA fragment.
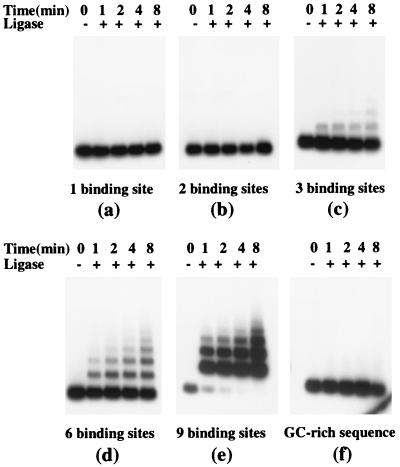
To determine whether the slowly migrating TnpA–DNA complexes are intermolecular, we investigated the ability of TnpA to accelerate the formation of DNA multimers in the presence of ligase (17, 21). Accelerated ligation in this assay requires the ends of different DNA molecules to be brought together actively. DNA fragments containing the TnpA binding sites and control DNA fragments lacking TnpA binding sites were incubated briefly with ligase in the presence and absence of TnpA. The ligation products then were deproteinized and analyzed on agarose gels (see Materials and Methods). DNA fragments containing either one or two TnpA binding sites did not form DNA multimers under these conditions, nor did DNA fragments lacking TnpA binding sites (Fig. 2 a, b, and f). By contrast, addition of TnpA accelerated ligation of fragments containing three or more TnpA binding sites (Fig. 2 c, d and e). The multimers are linear as judged by nuclease sensitivity (data not shown). Moreover, at identical DNA and ligase concentrations, the rate of multimerization accelerated markedly as the number of binding sites per DNA fragment increased. TnpA lacking the dimerization domain bound to DNA containing TnpA binding sites but did not accelerate ligation (data not shown). We infer that DNA-bound TnpA molecules interact, accelerating ligation by bringing DNA ends into close proximity.
Figure 2.
TnpA-mediated formation of DNA multimers in the presence of ligase. 32P-labeled DNA fragments (20 ng) containing (a–e) or lacking (f, the GC-rich region of Spm) TnpA binding sites were incubated with 120 ng of TnpA in the presence or absence of ligase for the time indicated, were deproteinized, were fractionated on a 1% agarose gel, and were detected by autoradiography.
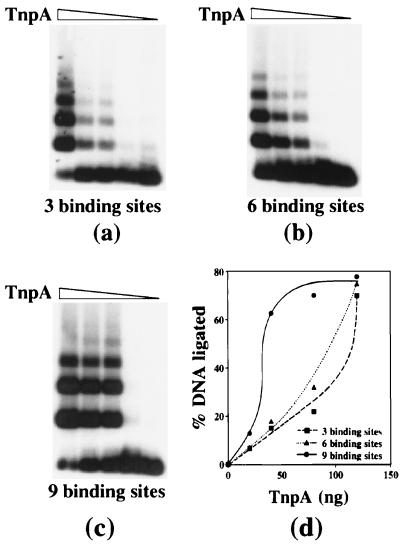
A ligation experiment was performed with DNA fragments containing 3, 6, and 9 binding sites at increasing TnpA concentrations. The fraction of ligated DNA increased over a significantly narrower range of TnpA concentrations for fragments containing nine binding sites than for fragments containing six and three binding sites, describing the sigmoidal saturation curve characteristic of cooperative interactions (Fig. 3).
Figure 3.
The effect of binding site number on TnpA-mediated DNA fragment ligation. (a–c) DNA fragments containing TnpA binding sites were ligated for 10 min in the presence of increasing amounts of TnpA, were fractionated on a 1% agarose gel, and were detected by autoradiography. (d) The percentage of shifted DNA in a–c is plotted as a function of the TnpA amount.
TnpA Interacts with TnpD.
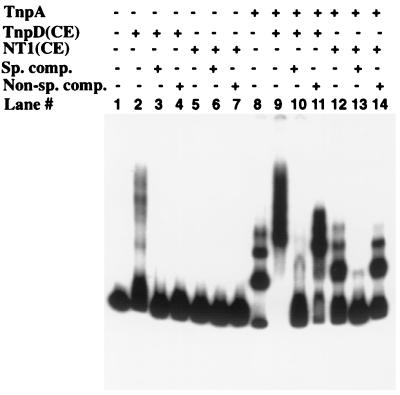
Both TnpA and TnpD, a second Spm-encoded protein, are required for transposition. Because TnpD has not been expressed yet in a heterologous system, we used extracts of transgenic tobacco cell lines expressing a tnpD cDNA from a CaMV 35S promoter. The TnpD-containing extract retarded the mobility of an oligonucleotide containing TnpA binding sites (Fig. 4, lane 2) whereas an extract of untransformed cells did not (Fig. 4, lane 5). The mobility shift was abolished by both specific and nonspecific competitor DNAs (Fig. 4, lanes 3 and 4). Similar results were obtained by using the complete Spm promoter fragment, confirming that TnpD alone binds DNA nonspecifically (data not shown). However, addition of a TnpD-containing extract supershifted a TnpA–DNA complex whereas addition of a comparable extract lacking TnpD had no effect on its mobility (Fig. 4, lanes 8, 9, and 12). Unlabeled competitor oligonucleotide containing TnpA binding sites abolished the supershift caused by TnpD as well as the TnpA-associated decrease in fragment mobility (Fig. 4, lane 10). However, a supershift still was observed in the presence of nonspecific competitor DNA (Fig. 4, compare lanes 11 and 14), implying that the supershift is attributable to the interaction of TnpD with TnpA.
Figure 4.
Binding of TnpD to DNA and DNA–TnpA complexes. A 279-bp DNA fragment containing two TnpA binding sites was used as the binding target DNA (Fig. 1b). Binding reactions were performed in DNA binding buffer and contained 80 ng of TnpA and 10 μg of protein from cell extracts. The extra, slow-migrating band in lanes 8 and 12 is likely the result of nonspecific interaction of proteins (BSA in lane 8 and NT1 cell extract in lane 12) with TnpA bound to DNA at the high protein concentrations used to equalize the total amount of protein. DNA–protein complexes were fractionated on a native 4% polyacrylamide gel. TnpA, E. coli over-expressed TnpA protein; TnpD(CE), extract of tobacco SR1 cells expressing TnpD; NT1(CE), extract of tobacco NT1 cells; Sp. comp., specific competitor DNA; Non-sp. comp., non-specific competitor DNA.
To determine whether TnpA and TnpD interact directly, we used a yeast two-hybrid system modified for use in plant cells (13, 22). We created the following two types of hybrid genes: GAL4/TnpD fusions (Fig. 5a) and TnpA/VP16 fusions (Fig. 5b). A firefly LUC reporter gene was constructed by using a minimal CaMV promoter and GAL4 DNA binding sites (Fig. 5c). Plasmids carrying the GAL4/TnpD, TnpA/VP16, and LUC constructs were introduced into cultured tobacco cells by particle bombardment together with an internal control plasmid carrying a chloramphenicol acetyl transferase gene expressed from a CaMV 35S promoter. LUC and chloramphenicol acetyl transferase activities were measured, and the results are expressed as their ratio (Fig. 5d).
A fusion protein comprising the N-terminal 120 amino acids of TnpA and the VP16 transcription activation domain interacts with the GAL4/TnpD fusion to activate expression of the LUC gene 6-fold over the level observed with the N terminus of TnpA alone (Fig. 5d, lanes 1 and 2). Inclusion of the TnpA DNA-binding domain (amino acids 120–422) in the TnpA/VP16 fusion yields similar results (Fig. 5d, lanes 3 and 4). Inclusion of the protein dimerization domain of TnpA (amino acids 422–543) substantially enhances the ability of the TnpA/VP16 fusion to interact with TnpD, as judged by the 30-fold activation of LUC expression (Fig. 5d, lanes 5 and 6), yielding a value within a factor of two of that observed with a GAL4/VP16 fusion (Fig. 5d, lane 17). The observation that the TnpD fragment expressed in this fusion protein lacks amino acids 1–110 implies that the N-terminal 110 amino acids of TnpD are not required for TnpD-TnpA interactions and that the fusion protein is stable. By contrast, GAL4/TnpD fusion proteins lacking either the N-terminal 367 amino acids or the C-terminal 12 amino acids of TnpD do not interact with TnpA/VP16 fusions to activate LUC gene expression (Fig. 5d, lanes 11–15). Additional controls show that LUC gene activation mediated by the VP16 activation domain and the GAL4 DNA-binding domain depends on the interaction between TnpA and TnpD (Fig. 5d, lanes 7–10). We conclude that TnpA and TnpD interact directly in vivo.
TnpD Stabilizes the TnpA–DNA Complex.
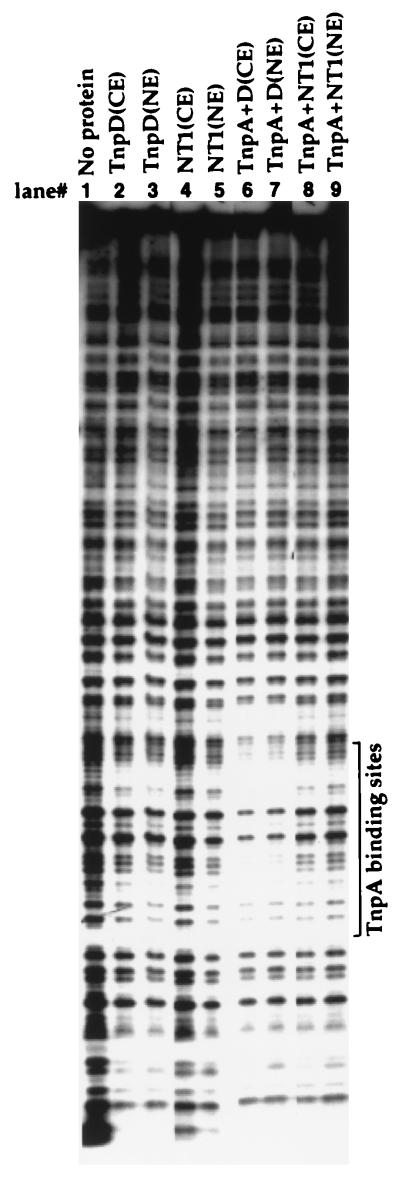
TnpA alone does not protect its binding site from DNase I digestion (Fig. 6, lanes 8 and 9), as reported (4). However, a DNase I footprint is seen when the TnpA-binding site oligonucleotide is preincubated with both TnpA and tobacco cell or nuclear extract containing TnpD (Fig. 6, lanes 6 and 7). No footprint is observed with extracts of either TnpD-expressing or untransformed cells in the absence of TnpA (Fig. 6, lanes 2–5), nor were footprints observed with extracts of untransformed cells in the presence of TnpA alone (Fig. 6, lanes 8 and 9). Similar experiments using a 5′ terminal Spm fragment showed protection of the entire sequence but only when both TnpA and TnpD were present (data not shown). Because the oligonucleotide sequence protected from DNase I digestion is the TnpA binding site, we attribute the footprint to stabilization of the TnpA–DNA complex by the binding of TnpD to TnpA.
Figure 6.
DNase I footprint analysis of protein–DNA complexes. A labeled DNA fragment containing a tail-to-tail dimeric TnpA binding motif was incubated with the proteins indicated at the top of each lane before digestion with DNase I. The digested products were fractionated on a denaturing 8% polyacrylamide gel. TnpD (CE) and TnpD(NE) were whole cell and nuclear extracts, respectively, of transgenic tobacco cells expressing TnpD from CaMV 35S promoter; NT1(CE) and NT1(NE) were whole cell and nuclear extracts, respectively, of untransformed tobacco cells. The position of TnpA binding sites was determined by Maxam–Gilbert sequencing of the labeled DNA.
DISCUSSION
Transposition and TnpA Binding Sites at Spm Termini.
The 5′ and 3′ ends of the Spm element have 9 and 15 TnpA-binding motifs, respectively, defined as sequences with 75% or more identity to the consensus sequence CCGACACTCTTA (7). It has been suggested that only 14 of these are TnpA binding sites, based on the observation that a G residue in this subset is hypermethylated by dimethyl sulfate in the presence of TnpA (4). However, the methylated G residue in 13 of the 14 methylated binding sites occurs in the submotif CCGA, the 14th being CCGT. Of the remaining 10 sites, 5 lack a methylatable G residue at the corresponding position and 3 have other sequence variations in the submotif. Two of the binding sites in the three-binding-site fragment used here contain the CCGA submotif whereas the third contains a CCCA submotif in the corresponding position. All three bind TnpA efficiently, suggesting that most or all of the sites identified by sequence similarity to the consensus TnpA binding sequence are true binding sites.
Intraelement deletions that extend into the subterminal regions of the Spm transposon at both 5′ and 3′ ends have a major effect on transposition, delaying the timing in development and reducing the frequency, even in the presence of a transposase source. However, a deletion of the 5′ subterminal GC-rich sequence of the transposon is epistatic in its effect on transposition to deletions that remove TnpA binding sites, making it difficult to evaluate the requirement for multiple binding sites at the 5′ end (7, 23, 24). Interpretation of deletions at the 3′ end of the element is more straightforward. McClintock’s mutants dSpm-7977B and dSpm-7995 lack the first four and five TnpA binding sites just distal to the transcription termination site, respectively, but show unchanged transposition frequencies with respect to the parent element (7). A more extensive deletion derivative, designated CS12, which retains the six TnpA binding sites closest to the 3′ end of the element (G. Bunkers, S.G. Pickett, and V. Raboy, personal communication), transposes at a very low frequency and very late in development (23, 25). Thus, 10 binding sites suffice for normal transposition, but 6 do not, an observation in good agreement with results of the present TnpA binding studies.
DNA Binding Properties of TnpA.
The binding properties of TnpA contrast markedly with the highly cooperative binding observed with many DNA binding proteins that have multiple tandem binding sites (26–29). Although the 5′ and 3′ ends of the Spm transposon contain 9 and 15 TnpA binding sites, respectively, the initial binding of the protein to DNA does not exhibit cooperativity, and it appears likely from the present data that the protein binds as a monomer. Although its initial binding is relatively unaffected by the number of binding sites per DNA fragment, intermolecular DNA–protein complexes form with DNA fragments containing three or more TnpA binding sites. The efficiency of complex formation increases dramatically as the number of TnpA binding sites per DNA molecule doubles and triples. Moreover, intermolecular complex formation exhibits cooperativity with respect to TnpA concentration. Thus, TnpA binding site redundancy appears to underlie and drive the efficient formation of transposition complexes containing Spm termini crosslinked by TnpA molecules.
TnpD Stabilizes TnpA–DNA Complexes.
There is evidence of several kinds that TnpD interacts directly with TnpA. The ability of TnpD-containing plant extracts to supershift TnpA–DNA complexes and to stabilize them sufficiently to protect the TnpA binding site from DNase I digestion suggests that TnpD may be a component of the transposition complex as well, increasing the affinity of TnpA for or its residence time on binding site-containing DNA. Indeed, we interpret the observation that a TnpA binding site footprint is detectable only in the presence of both TnpA and TnpD as additional evidence that TnpA alone does not bind tightly to its binding site.
In two-hybrid experiments with TnpA/VP16 and GAL4/TnpD fusion proteins, the relationship between free and DNA-bound Spm proteins is reversed. That is, the DNA-binding component is provided by the binding domain of the GAL4 protein rather than that of TnpA. The observation that the TnpA/VP16 fusion protein was able to interact with the GAL4/TnpD fusion protein, as measured by activation of reporter gene expression, implies that TnpD can interact directly with TnpA and not just with DNA-bound TnpA. Moreover, the activation of the reporter gene was significantly higher with TnpA derivatives containing the protein dimerization domain than with derivatives lacking it, suggesting that TnpD may interact with more than one TnpA molecule.
The implication of the present experiments is that the developmental timing and frequency of transposition are determined not only by the concentration of the element-encoded proteins necessary for transposition but also by the number of TnpA binding sites at element ends. The high multiplicity of binding sites serves to stabilize TnpA–DNA complexes, as does the interaction between TnpA and TnpD. The net effect of these interactions is to facilitate formation of a protein-crosslinked transposition complex at low protein concentrations. However, there is nothing inherent in TnpA crosslinking of DNA molecules to align element ends. If, as is likely, alignment of termini is important in transposition, then the ability of TnpA to slide along the DNA may be important in forming a final transposition complex in which the element ends are in close juxtaposition. Our present working hypothesis is that there is a highly specific interaction between TnpA, TnpD, and element ends to align them for transposition. In view of the substantially greater abundance of TnpA than TnpD, it may be that TnpD does not reach a concentration sufficient to interact with the transposition complex until most ends have been brought together by TnpA crosslinking. TnpD may then nucleate alignment of ends by preferentially interacting with TnpA molecules adjacent to the terminal inverted repeats to cleave (or position the cleavage of) element termini. By mechanisms not yet understood, target insertion sites are recruited to the transposition complex, perhaps together with additional proteins not encoded by the element, initiating the cleavage, strand exchange, ligation, and fill-in reactions required for transposition.
Acknowledgments
We thank Mary Strem and Zhijian Chen for technical assistance, Dhruba K. Chattoraj for advice on ligation kinetics experiments, and Jill Deikman and Ryuji Tsugeki for valuable comments on the manuscript. This research was supported by National Institutes of Health Grant GM34296. M.S. was supported by Advanced Researcher Grant 84FI-037132 from the Swiss National Science Foundation and by the Carnegie Institution of Washington.
ABBREVIATIONS
- Spm
Suppressor–mutator
- LUC
luciferase
- CaMV
cauliflower, mosaic virus
References
- 1.Frey M, Reinecke J, Grant S, Saedler H, Gierl A. EMBO J. 1990;9:4037–4044. doi: 10.1002/j.1460-2075.1990.tb07625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masson P, Strem M, Fedoroff N. Plant Cell. 1991;3:73–85. doi: 10.1105/tpc.3.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masson P, Rutherford G, Banks J A, Fedoroff N. Cell. 1989;58:755–765. doi: 10.1016/0092-8674(89)90109-8. [DOI] [PubMed] [Google Scholar]
- 4.Gierl A, Lutticke S, Saedler H. EMBO J. 1988;7:4045–4053. doi: 10.1002/j.1460-2075.1988.tb03298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gierl A, Schwarz-Sommer Z, Saedler H. EMBO J. 1985;4:579–583. doi: 10.1002/j.1460-2075.1985.tb03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. EMBO J. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masson P, Surosky R, Kingsbury J, Fedoroff N V. Genetics. 1987;117:117–137. doi: 10.1093/genetics/117.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClintock B. Year Book—Carnegie Inst Washington. 1955;54:245–255. [PubMed] [Google Scholar]
- 9.McClintock B. Year Book—Carnegie Inst Washington. 1957;56:393–401. [PubMed] [Google Scholar]
- 10.McClintock B. Brookhaven Symp Biol. 1965;18:162–184. [PubMed] [Google Scholar]
- 11.McClintock B. Year Book—Carnegie Inst Washington. 1971;70:5–17. [PubMed] [Google Scholar]
- 12.Trentmann S M, Saedler H, Gierl A. Mol Gen Genet. 1993;238:201–208. doi: 10.1007/BF00279548. [DOI] [PubMed] [Google Scholar]
- 13.Schläppi M, Raina R, Fedoroff N. Plant Mol Biol. 1996;32:717–725. doi: 10.1007/BF00020212. [DOI] [PubMed] [Google Scholar]
- 14.Schläppi M, Raina R, Fedoroff N. Cell. 1994;77:427–437. doi: 10.1016/0092-8674(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 15.Raina R, Cook D, Fedoroff N. Proc Natl Acad Sci USA. 1993;90:6355–6359. doi: 10.1073/pnas.90.13.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 17.Mukhopadhyay G, Sozhamannan S, Chattoraj D K. EMBO J. 1994;13:2089–2096. doi: 10.1002/j.1460-2075.1994.tb06484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell J A, Roy M K, Sanford J C. In Vitro Cell Dev Biol. 1992;28:97–105. [Google Scholar]
- 19.Foster R, Gasch A, Kay S, Chua N-H. In: Analysis of Protein/DNA Interactions. Koncz C, Chua N-H, Schell J, editors. Teaneck, NJ: World Scientific; 1993. pp. 378–392. [Google Scholar]
- 20.Masson P, Fedoroff N V. Proc Natl Acad Sci USA. 1989;86:2219–2223. doi: 10.1073/pnas.86.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miron A, Mukherjee S, Bastia D. EMBO J. 1992;11:1205–1216. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unger E, Parsons R L, Schmidt R J, Bowen B, Roth B A. Plant Cell. 1993;5:831–841. doi: 10.1105/tpc.5.8.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiefelbein J W, Furtek D B, Dooner H K, Nelson O., Jr Genetics. 1988;120:767–677. doi: 10.1093/genetics/120.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tacke E, Z, Schwarz-Sommer Z, Peterson P W, Saedler H. Maydica. 1986;31:83–91. [Google Scholar]
- 25.Schiefelbein J W, Raboy V, Fedoroff N, Nelson O E., Jr Proc Natl Acad Sci USA. 1985;82:4783–4787. doi: 10.1073/pnas.82.14.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers R M, Rio D C, Robbins A K, Tjian R. Cell. 1981;25:373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- 27.Desplan C, Theis J, O’Farrell P H. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss V, Claverie-Martin F, Magasanik B. Proc Natl Acad Sci USA. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J D, Pirrotta V. EMBO J. 1993;12:2075–2083. doi: 10.1002/j.1460-2075.1993.tb05856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]