Abstract
Caspase-mediated proteolysis is a critical and central element of the apoptotic process; therefore, it is important to identify the downstream molecular targets of caspases. We established a method for cloning the genes of caspase substrates by two major modifications of the yeast two-hybrid system: (i) both large and small subunits of active caspases were expressed in yeast under ADH1 promoters and the small subunit was fused to the LexA DNA-binding domain; and (ii) a point mutation was introduced that substituted serine for the active site cysteine and thereby prevented proteolytic cleavage of the substrates, possibly stabilizing the enzyme–substrate complexes in yeast. After screening a mouse embryo cDNA expression library by using the bait plasmid for caspase-3, we obtained 13 clones that encoded proteins binding to caspase-3, and showed that 10 clones including gelsolin, an actin-regulatory protein implicated in apoptosis, were cleaved by recombinant caspase-3 in vitro. Using the same bait, we also isolated human gelsolin cDNA from a human thymus cDNA expression library. We showed that human gelsolin was cleaved during Fas-mediated apoptosis in vivo and that the caspase-3 cleavage site of human gelsolin was at D352 of DQTD352G, findings consistent with previous observations on murine gelsolin. In addition, we ascribed the antiapoptotic activity of gelsolin (which we previously reported) to prevention of a step leading to cytochrome c release from the mitochondria into the cytosol. Our results indicate that this cloning method is useful for identification of the substrates of caspases and possibly also of other enzymes.
Apoptosis is a fundamental cellular process involved in a variety of biological phenomena, including morphogenesis and maintenance of tissue homeostasis. ced-3 is a gene of the nematode Caenorhabditis elegans that is required for cell death and it encodes a protein resembling interleukin-1β-converting enzyme, a cysteine protease that cleaves the inactive 31-kDa precursor of interleukin-1β to generate the active cytokine (1, 2). At least 10 proteins homologous to interleukin-1β-converting enzyme and CED-3 have been identified in mammals and are classified into three subfamilies (caspase-1-, caspase-2-, and caspase-3-like proteases) based on their structures (3). Caspases are synthesized as dormant proenzymes and are proteolytically activated in response to various death signals (4). The active form is composed of two subunits, large and small, both of which are derived from the proenzyme by cleavage at the C-terminal side of specific Asp residues (1–4). The caspases have been suggested to constitute a protease cascade (4–7).
Because caspase-mediated proteolysis is a critical and central element of the apoptotic process (8–10), identification of the crucial downstream molecular targets of these proteases is inevitable for understanding apoptotic signal transduction. Various structural and signaling proteins have been shown to be cleaved by caspases during apoptotic cell death (8, 9) including ICAD, an inhibitor of caspase-activated DNase, which is essential for internucleosomal DNA degradation but not for the execution of apoptosis (11, 12). Gelsolin, an actin-regulatory protein that modulates cytoplasmic actin gel–sol transformation (13), is implicated in apoptosis on the basis of (i) its cleavage during apoptosis in vivo (14); (ii) its cleavage by caspase-3 in vitro (14); (iii) prevention of apoptosis by its overexpression (15); and (iv) induction of apoptosis by one of the cleaved products (14). Gelsolin has Ca2+-activated multiple activities, severs actin filaments, and caps the fast growing ends of the filaments, and also nucleates actin polymerization (16–18).
In this study, we developed a systematic method of cloning genes for the substrates of caspases by modification of the yeast two-hybrid system and identified gelsolin as a substrate of caspase-3.
MATERIALS AND METHODS
Plasmid Construction for the Yeast Two-Hybrid System.
PCR was used to construct expression plasmids in yeast. The PCR primers employed for human caspase-1 were p20–5′ (5′-CGGGATCCACCATGAACCCAGCTATGCCC-3′) and p20–3′ (5′-CGGAATTCTTAATCTTTAAACCACAC-3′) corresponding to the N-terminal and C-terminal regions of the p20 subunit of caspase-1 cDNA, as well as p10–5′ (5′-CGGAATTCGCTATTAAGAAAGCC-3′) and p10–3′ (5′-ACGGATCCCTTAATGTCCTGGGAA-3′) corresponding to the N-terminal and C-terminal regions of the p10 subunit of caspase-1 cDNA. For human caspase-3, the primers were p17–5′ (5′-CGGATATCACCATGTCTGGAATATCCCTG3′) and p17–3′ (5′-CGGATATCTTAGTCTGTCTCAATGCC-3′) corresponding to the N-terminal and C-terminal regions of the p17 subunit of caspase-3 cDNA, as well as p12–5′ (5′-CGGAATTCAGTGGTGTTGATGAT-3′) and p12–3′ (5′-CGGGATCCTTAGTGATAAAAATAGAG-3′) corresponding to the N-terminal and C-terminal regions of the p12 subunit of caspase-3 cDNA. The primers for crmA were crmA-5′ (5′-CGCTCGAGGAATGGATATCTTCAGG-3′) and crmA-3′ (5′-CGGAATTCTTAATTAGTTGTTGG-3′) corresponding to the N-terminal and C-terminal regions of crmA cDNA. p20–5′ and p17–5′ primers have BamHI and EcoRV sites with a Kozak sequence; p20–3′ and p17–3′ primers have EcoRI and EcoRV sites with stop codon; p10–5′, p12–5′, and crmA-3′ primers have an EcoRI site; p10–3′ and p12–3′ primers have a BamHI site with stop codon; and crmA-5′ primer has an XhoI site at the 5′ end. The amplified human caspase-1, caspase-3, and crmA fragments were cloned into pBluescript SK(−) (Stratagene) and sequenced.
A fragment encoding caspase-1-p20 and caspase-1-p10 was generated by PCR by using caspase-1 cDNA bearing an active site mutation (Cys-285 to Gly) as the template, and was cloned into the BamHI–EcoRI site of pGAD10 and the EcoRI–BamHI site of pBTM116 to generate pGAD-casp1-p20m and pBTM-casp1-p10, respectively. The BamHI–EcoRI fragment encoding caspase-1-p20 was blunt-ended and cloned into the blunt-ended HindIII site of pGAD10 lacking the Gal4 activation domain to generate pGADΔAD-casp1-p20m. The SnaBI–EcoRV fragment bearing caspase-1-p20 under control of the ADH1 promoter from pGADΔAD-casp1-p20m was cloned into the PvuII site of pBTM-casp1-p10 to generate pBTM-casp1-p10p20m.
A fragment encoding caspase-3-p17 and caspase-3-p12 was generated by PCR by using caspase-3 cDNA bearing an active site mutation (Cys-163 to Ser) (19, 20) as template, and was cloned into the EcoRV site of pGAD10 and the EcoRI–BamHI site of pBTM116 to generate pGAD-casp3-p17m and pBTM-casp3-p12, respectively. The EcoRV fragment encoding caspase-3-p17 was cloned into the blunt-ended HindIII site of pGAD10 lacking the Gal4 activation domain to generate pGADΔAD-casp3-p17m. The SnaBI–EcoRV fragment bearing caspase-3-p17 under control of the ADH1 promoter from pGADΔAD-casp3-p17m was cloned into the PvuII site of pBTM-casp3-p12 to generate pBTM-casp3-p12p17m.
A fragment encoding crmA was generated by PCR by using the p996 plasmid (21) as template, and was cloned into the XhoI–EcoRI site of pGAD10 to generate pGAD-crmA.
Yeast Two-Hybrid Screening.
Yeast two-hybrid screening using pBTM-casp3-p12p17m as the bait was performed with mouse 11-day embryo and human thymus cDNA expression libraries fused to the Gal4 activation domain in the pGAD10 plasmid following the matchmaker Two-Hybrid System Protocol (CLONTECH) in L40 cells (MATa trp1 leu2 his3 ade2 LYS2∷lexA-HIS3 URA3∷lexA-lacZ).
In Vitro Cleavage by Recombinant Caspase-3.
To construct expression plasmids from positive clones obtained from the yeast two-hybrid screening, the EcoRI fragments of positive clones were inserted into the EcoRI site of pRSET B (Invitrogen), in which the sequences were under control of the T7 promoter and joined in-frame to sequences encoding an N-terminal fusion peptide with an ATG translation initiation codon and the sequence for six successive histidine residues. [35S]Methionine-labeled proteins were prepared by using expression plasmids and a TnT T7-coupled in vitro transcription and translation system (Promega) according to the manufacturer’s instructions, and were subjected to cleavage by recombinant caspase-3, which was prepared as described (19, 20). Recombinant caspase-3 was preincubated at room temperature for 15 min with or without 1 μM Ac-DEVD-CHO (Peptide Institute, Mino, Japan) in 25 μl of a buffer [50 mM Hepes/0.1 M NaCl/10% (vol/vol) glycerol, pH 7.5/10 mM DTT], and the reaction was initiated by addition of 5 μl of [35S]methionine-labeled protein. After incubation at 37°C for 2 hr, the reaction was stopped by addition of SDS/PAGE sample buffer, and cleaved products were analyzed by SDS/12% PAGE.
Cell Culture and Induction of Apoptosis.
HepG2 (a human hepatocellular carcinoma cell line) and Jurkat (a lymphoblastoid T-cell line) cells were cultured in RPMI 1640 medium with 10% fetal bovine serum. JGF-5 and JGF-7 are derivatives of Jurkat expressing gelsolin, and JNF-2 is a control transfectant (15). For induction of apoptosis, HepG2 cells were treated with 1 μg/ml of an agonistic anti-Fas antibody (CH-11; Medical and Biological Laboratories, Nagoya, Japan) in the presence of 0.2 μg/ml actinomycin D, and Jurkat, JGF-5, JGF-7, and JNF-2 cells were treated with 1 μg/ml of the agonistic anti-Fas antibody, for various periods.
Antibodies and Western Blot Analysis.
Anti-caspase-3 mAb for detection of procaspase-3, anti-caspase-3 polyclonal antibodies for detection of caspase-3-p12, anti-gelsolin mAb, and anti-cytochrome c antibody were purchased from Transduction Laboratories (Lexington, KY), Santa Cruz Biotechnology, Sigma, and PharMingen, respectively. Western blot analysis was carried out essentially as described (22).
Construction and Purification of Human Cytoplasmic Gelsolin.
PCR was used to construct expression plasmid of human cytoplasmic gelsolin in Escherichia coli. The PCR primers employed were 5′-NdeI (5′-GCGCGGCCCAGCCATATGGTGGTGGAG-3′) and A10 (5′-CCGCCCTTTGACCTGGAA-3′). 5′-NdeI primer has a NdeI site with an initiation codon at the 5′ end. A fragment encoding the 1–144 amino acid residue of human gelsolin was generated by PCR by using LKCG (23) plasmid as the template and sequenced. The amplified fragment digested with NdeI and NotI and the NotI–BamHI fragment from LKCG plasmid were ligated and cloned into the NdeI–BamHI site of pET-11a (Novagen) to generate pET-HCG. Purification of human cytoplasmic gelsolin expressed in E. coli was carried out essentially as described (24).
Cytochrome c Release Assay.
Cells were harvested by centrifugation at 600 × g for 5 min at 4°C, and the cell pellets were suspended in buffer (20 mM Hepes, pH 7.4/10 mM KCl/1.5 mM MgCl2/1 mM EDTA/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride/250 mM sucrose). The cells were treated with 0.3 mg/ml digitonin for 3 min at 37°C and centrifuged at 12,000 × g for 5 min at 4°C. The pellets and supernatants were respectively used for Western blot analysis as the membrane fraction including heavy membranes (containing mitochondria) and nuclei, and the cytosolic fraction also containing light membranes.
RESULTS
Strategy for Molecular Cloning of Genes for Caspase Substrates.
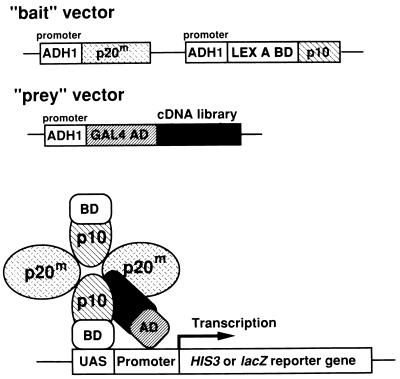
In our cloning method for genes encoding caspase substrates using the yeast two-hybrid system (Fig. 1), both large and small subunits of active caspases were separately expressed in yeast under ADH1 promoters to intentionally maintain an equimolar ratio of large-to-small subunits. Because the N-terminal parts of the small subunits in the active forms of caspase-1 and caspase-3 seem to face the outside of the complexes, judging from their three-dimensional crystal structures (25–27), the small subunit was fused to the LexA DNA-binding domain. Furthermore, to possibly stabilize the enzyme–substrate complex in yeast, a point mutation was introduced that substituted serine for the active site cysteine and thereby prevented proteolytic cleavage of the substrates. To confirm the feasibility of this method, we first examined the interaction of the large subunit with the small subunit of caspase-1 in yeast two-hybrid assays. Caspase-1-p10 was able to bind to caspase-1-p20, but not CrmA (Table 1). In addition, cotransfection of pBTM-casp1-p10p20m, which expressed both caspase-1-p10 and caspase-1-p20 under ADH1 promoters, and pGAD-crmA (CrmA is a good caspase-1 substrate) (29) yielded His+ transformants that were also β-galactosidase positive (Table 1), suggesting that a stable complex of caspase-1 and CrmA was formed in yeast. These results indicated that our strategy of identifying genes for caspase substrates using the two-hybrid system was feasible.
Figure 1.
Diagram of the method of cloning genes of caspase targets using the modified yeast two-hybrid system. The bait vector contained sequences for both the large (p20) and small (p10) subunits of active caspase-1, which were expressed separately in yeast under ADH1 promoters, and the small subunit was fused to the LexA DNA-binding domain. A point mutation was introduced into the active site cysteine to prevent proteolytic cleavage of the substrates. When the bait and prey vectors were cotransfected into yeast a stable enzyme–substrate complex was formed in yeast.
Table 1.
Complex formation by large and small caspase subunits with substrate
| pBTM (LexA DB)- | pGAD (Gal4 AD)- | β-Galactosidase activity |
|---|---|---|
| casp1-p10 | casp1-p20m | + |
| casp1-p10 | RAD51 | − |
| RAD51 | casp1-p20m | − |
| RAD51 | RAD51 | ++ |
| casp1-p10 | CrmA | − |
| RAD51 | CrmA | − |
| casp1-p10p20m | CrmA | + |
| casp1-p10p20m | RAD51 | − |
Yeast L40 cells were cotransformed with expression plasmids for LexA DNA-binding domain fusion proteins and for Gal4 activation domain fusion proteins as indicated. Each transformation mixture was plated on a synthetic dropout plate lacking leucine, tryptophan, and histidine. Filter assays for β-galactosidase activity were performed to detect interactions between fusion proteins. Rad51 (28) was used as a control. ++ and +, development of blue color within 1 hr and 2 hr, respectively; −, no growth of transformed yeast colonies.
Cloning of Genes for Caspase-3 Substrates.
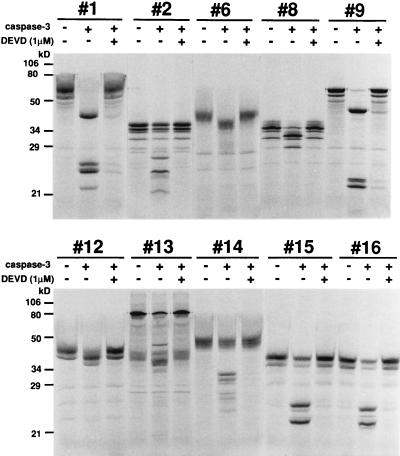
Because caspase-3 has been implicated as a key mediator of apoptosis in mammalian cells (30), we also constructed a bait plasmid for caspase-3, pBTM-casp3-p12p17m, as described in Materials and Methods. This bait plasmid was cotransfected into yeast with prey plasmids containing a mouse 11-day embryo cDNA expression library fused to the Gal4 activation domain. Sixty-nine positive clones were obtained by screening 4.2 × 107 transformants. From the size of the inserted fragments, the restriction enzyme digestion pattern, and partial DNA sequencing, the 69 clones were divided into 13 groups. To examine cleavage of the candidates by caspase-3 in vitro, proteins encoded by the cDNAs of the positive clones were produced as described. As shown in Fig. 2, 10 of 13 candidates were cleaved by recombinant caspase-3 and cleavage was inhibited in the presence of 1 μM Ac-DEVD-CHO, suggesting that these proteins were possible substrates for caspase-3. DNA sequencing analysis showed that three of the positive clones (clones #1, #9, and #12) encoded overlapping parts of mouse gelsolin, which is an actin-regulatory protein and was recently found to be cleaved by caspase-3 (14). We analyzed further these gelsolin clones in this study, and the other clones will be described elsewhere. Clones #1, #9, and #12 contained residues 197–731, 234–731, and 9–415 of mouse gelsolin, respectively. Screening of a human thymus cDNA expression library by using the same bait (pBTM-casp3-p12p17m) resulted in isolation of two clones encoding human gelsolin from residues 202–731 and 276–731. These results indicated that our method could successfully identify caspase substrates.
Figure 2.
In vitro cleavage of candidate proteins by recombinant active caspase-3. [35S]Methionine-labeled proteins from 10 positive clones and 0.6 μg of purified recombinant caspase-3 were incubated with or without 1 μM Ac-DEVD-CHO for 2 hr, and the reaction products were analyzed by SDS/10% PAGE.
Cleavage of Human Gelsolin by Caspase-3.
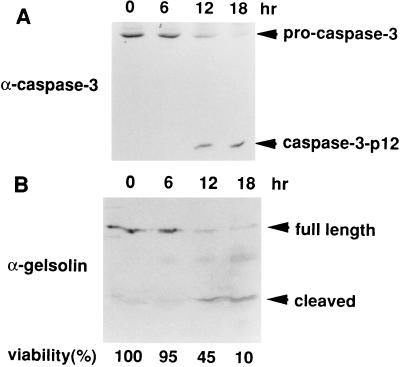
To examine the cleavage of gelsolin during apoptosis in vivo, HepG2 cells were treated with an agonistic anti-Fas antibody. As shown in Fig. 3, gelsolin was cleaved with similar kinetics to those of caspase-3 during apoptosis, suggesting that it was directly cleaved by caspase-3 in vivo.
Figure 3.
In vivo cleavage of gelsolin during Fas-mediated apoptosis of HepG2 cells. HepG2 cells were treated with an agonistic anti-Fas antibody for the indicated periods. Cells were harvested, the lysates were subjected to SDS/15% PAGE and were immunoblotted with anti-caspase-3 antibodies (A) and anti-gelsolin antibody (B). Cell viability (%) was determined by morphological analysis after staining with Hoechst 33342 and propidium iodide (31), and is indicated at the bottom.
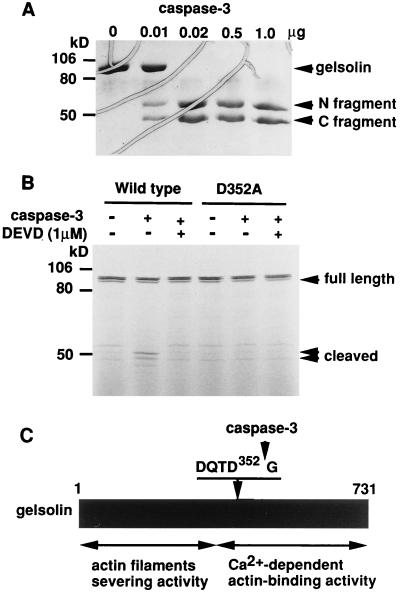
Bacterially expressed human gelsolin was purified and incubated with various amounts of purified recombinant caspase-3. The gelsolin was found to be cleaved by caspase-3 in an enzyme concentration-dependent manner (Fig. 4A). Peptide sequence analysis of the products after gelsolin was cleaved by recombinant caspase-3 revealed that caspase-3 cleaved gelsolin at D352 of DQTD352G, which fitted the consensus caspase-3 cleavage sequence (DXXD). A mutant gelsolin with Ala at Asp-352 was not cleaved by recombinant active caspase-3 (Fig. 4B). Thus, the caspase-3 cleavage site of human gelsolin was determined to be at the C terminal of Asp-352 (Fig. 4C), consistent with the cleavage site of mouse gelsolin (14).
Figure 4.
Determination of the cleavage site of human gelsolin. (A) In vitro cleavage of gelsolin by recombinant caspase-3. Purified human gelsolin (4 μg) was incubated with the indicated amounts of purified recombinant caspase-3 at 37°C for 2 hr and reaction products were analyzed by SDS/10% PAGE and stained with Coomassie blue. Bands corresponding to the N-terminal and C-terminal fragments are indicated. (B) Mutant human gelsolin D352A resists cleavage by caspase-3. [35S]Methionine-labeled wild-type and mutant (D352A) gelsolin were incubated with 0.6 μg of purified recombinant caspase-3 as described in A. The cleavage products were subjected to SDS/10% PAGE and visualized by autoradiography. (C) Diagram showing caspase-3-mediated cleavage of human gelsolin. The cleavage site is at the C-terminal side of Asp-352 and splits gelsolin into an N-terminal fragment with actin filament-cutting activity and a C-terminal fragment with Ca2+-dependent actin-binding activity.
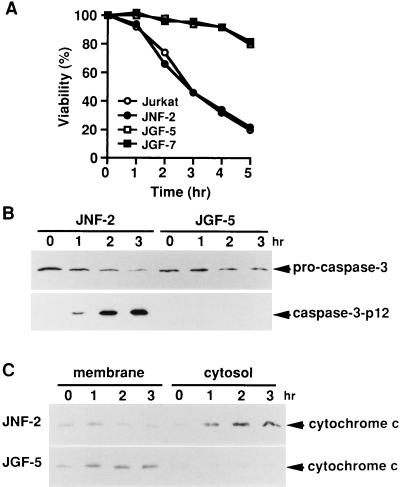
Fas-Induced Cytochrome c Release Is Inhibited by Overexpression of Gelsolin.
It was recently reported that overexpression of gelsolin prevents apoptosis induced by anti-Fas antibody, C2-ceramide, or dexamethasone through inhibition of caspase-3 activation (15). To study further the functional site(s) of gelsolin involved in preventing apoptosis, we examined the effect of gelsolin overexpression on the release of cytochrome c, which is a key event in apoptotic signal transduction (32–34). Once released into the cytoplasm by a still unknown mechanism, cytochrome c activates caspase-9 in concert with cytoplasmic factors dATP (or ATP) and Apaf-1 (a mammalian Ced-4 homolog), and subsequently activates caspase-3 (35). Treatment with the agonistic anti-Fas antibody induced apoptosis (Fig. 5A) and activation of caspase-3 (Fig. 5B) in control JNF-2 cells, whereas neither apoptosis nor caspase-3 activation was detected in gelsolin-overexpressing JGF-5 cells (Fig. 5 A and B), findings consistent with previous observations (15). Without anti-Fas antibody treatment, most of the detectable cytochrome c in both cell lines was in the membrane fraction containing mitochondria, whereas in JNF-2 cells treated with the anti-Fas antibody, cytochrome c was increased in the cytosol and correspondingly decreased in the membrane fraction (Fig. 5C). Similar results were obtained with parental Jurkat cells (data not shown). In contrast, there was little change of cytochrome c in the cytosolic and membrane fractions of JGF-5 cells (Fig. 5C) and JGF-7 cells (data not shown), indicating that overexpression of gelsolin prevented a step leading to cytochrome c release.
Figure 5.
Inhibition of cytochrome c release from the mitochondria into the cytoplasm by overexpression of gelsolin. (A) Viability of parental Jurkat cells, a vector-transfected derivative (JNF-2), and derivatives overexpressing gelsolin (JGF-5 and JGF-7) treated with anti-Fas antibody. After plating at a density of 1 × 104 cells per well in 96-well dishes, the cells were treated with an anti-Fas antibody for the indicated periods and viability was assessed morphologically by counting normal round cells as alive and fragmented cells as dead. Three independent experiments were carried out and more than 200 cells were counted in each experiment. (B) Processing of procaspase-3 after Fas-stimulation. Cells were treated with anti-Fas antibody for the indicated periods. Cell lysates (4 × 105 cells per lane) were fractionated by SDS/15% PAGE and immunoblotted with anti-caspase-3 antibodies. (C) Immunoblot analysis of cytochrome c in the membrane and cytosolic fractions. JNF-2 and JGF-5 cells were treated with anti-Fas antibody for the indicated periods, and membrane and cytosolic fractions were prepared as described. Each fraction (equivalent to 4 × 105 cells) was subjected to SDS/15% PAGE and immunoblotting with anti-cytochrome c antibody.
DISCUSSION
We have established a method for cloning the genes of caspase substrates by using the yeast two-hybrid system and obtained several candidate substrates for caspase-3, including gelsolin, that were cleaved by recombinant caspase-3 in vitro.
Gelsolin is an actin-regulatory protein that modulates actin gel-sol transformation by directly acting on actin filaments. We have shown that human gelsolin is cleaved by recombinant active caspase-3 at D352 of DQTD352G, which is known to be a caspase-3 cleavage site of murine gelsolin, and that a gelsolin with Ala instead of Asp at position 352 was not cleaved by caspase-3. The cleavage site was located between the N-terminal fragment that depolymerizes the actin cytoskeleton and the C-terminal fragment that binds actin in a Ca2+-dependent manner (Fig. 4C), consistent with the observation that caspase-cleaved gelsolin gains the ability to cut actin filaments in a Ca2+-independent manner (14). This study also confirmed the previous observation that gelsolin is cleaved during apoptosis (14).
Although gelsolin-deficient mice did not reveal any apoptosis-related abnormalities (36), direct involvement of gelsolin in apoptosis has been suggested by the recent observations from two groups. One group (15) in collaboration with ours has shown that gelsolin overexpression prevents apoptosis induced by anti-Fas antibody, C2-ceramide, or dexamethasone by inhibiting caspase-3 activation. The other group (14) has reported that caspase-cleaved gelsolin with the Ca2+-independent actin filament cleavage activity, can cause apoptotic cell death. Thus, gelsolin seems to have dual functions, i.e., it both prevents and, once cleaved, induces cell death. Similar dual functions have also been suggested for Bcl-2 (37).
The Fas-triggered death signal either directly activates the caspase cascade (type I) or acts indirectly via the mitochondria (type II), depending on the cell type (38). By using Jurkat cells (type II), we demonstrated that overexpression of gelsolin prevented Fas-induced cytochrome c release, suggesting that gelsolin may prevent apoptosis by acting upstream of the cytochrome c release step. Although there seems to be many signaling molecules involved in Fas-mediated apoptosis, such as FADD/MORT-1 (39, 40), p21-activated kinase 2 (PAK2) (41), Ras and Rac1 (42), JNK/SAPK and p38 kinase (42–44), Daxx (45), MKK6 (46), and caspases (5, 19, 31, 47–52), it is unclear which molecules are responsible for the release of cytochrome c. Because the sequence of the caspase-3 cleavage site of gelsolin (DQTD352G) resembles that of the baculovirus-encoded broad caspase inhibitor p35 (DQMDG) (53, 54), gelsolin itself might be a direct inhibitor of not only caspase-3 but also other caspases that may function upstream of the mitochondria. Alternatively, gelsolin may inhibit one or more steps leading to cytochrome c release by interacting with signaling molecules other than caspases. Thus, identification of the target(s) of gelsolin should make a contribution to better understanding of the apoptotic machinery.
Although the yeast two-hybrid system has already proven very useful for identifying interacting molecules, this, to the best of our knowledge, is the first successful application of the yeast system for identification of enzyme substrates. Our results suggest that a similar approach could be taken for additional enzymes, including proteases other than caspases, and kinases.
Acknowledgments
We thank Dr. Vishva M. Dixit for the pcDNA3/Yama plasmid, Dr. David J. Pickup for the p996 plasmid containing the crmA gene, Dr. David J. Kwiatkowski for the LKCG plasmid, and Dr. Akira Shinohara for pBTM116-RAD51 and pGAD10-RAD51. We also thank T. Ohtsubo for help with the construction of caspase-1 bait DNA. This work was supported in part by grants-in-aid for Scientific Research on Priority Areas and for Center of Excellence Research from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weigner J R, Aunins J, et al. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 2.Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Martin S J, Green D R. Cell. 1995;82:349–352. doi: 10.1016/0092-8674(95)90422-0. [DOI] [PubMed] [Google Scholar]
- 5.Enari M, Talanian R V, Wong W W, Nagata S. Nature (London) 1996;380:723–726. doi: 10.1038/380723a0. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu S, Eguchi Y, Kamiike W, Matsuda H, Tsujimoto Y. Oncogene. 1996;12:2251–2257. [PubMed] [Google Scholar]
- 7.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson D W, Thornberry N A. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 9.Villa P, Kaufmann S H, Earnshaw W C. Trends Biochem Sci. 1997;22:388–393. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- 10.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 11.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. Nature (London) 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 12.Sakahira H, Enari M, Nagata S. Nature (London) 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 13.Yin H L, Stossel T P. Nature (London) 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 14.Kothakota S, Azuma T, Reinhard C, Kippel A, Tang J, Chu K, McGarry T J, Kirschner M W, Koths K, Kwiatkowski D J, Williams L T. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsu M, Sakai N, Fujita H, Kashiwagi M, Gasa S, Shimizu S, Eguchi Y, Tsujimoto Y, Sakiyama Y, Kobayashi K, Kuzumaki N. EMBO J. 1997;16:4650–4656. doi: 10.1093/emboj/16.15.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin H L, Stossel T P. J Biol Chem. 1980;255:9490–9493. [PubMed] [Google Scholar]
- 17.Kurth M, Bryan J. J Biol Chem. 1984;259:10895–10903. [PubMed] [Google Scholar]
- 18.Janmey P A, Stossel T P. Nature (London) 1987;352:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 19.Kamada S, Washida M, Hasegawa J-I, Kusano H, Funahashi Y, Tsujimoto Y. Oncogene. 1997;15:285–290. doi: 10.1038/sj.onc.1201192. [DOI] [PubMed] [Google Scholar]
- 20.Kamada S, Funahashi Y, Tsujimoto Y. Cell Death Differ. 1997;4:473–478. doi: 10.1038/sj.cdd.4400268. [DOI] [PubMed] [Google Scholar]
- 21.Pickup D J, Ink B S, Hu W, Ray C A, Koklik W K. Proc Natl Acad Sci USA. 1986;83:7698–7701. doi: 10.1073/pnas.83.20.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada S, Shimono A, Shinto Y, Tsujimura T, Takahashi T, Noda T, Kitamura Y, Kondoh H, Tsujimoto Y. Cancer Res. 1995;55:354–359. [PubMed] [Google Scholar]
- 23.Cunningham C C, Stossel T P, Kwiatkowski D J. Science. 1991;251:1233–1236. doi: 10.1126/science.1848726. [DOI] [PubMed] [Google Scholar]
- 24.Fujita H, Laham L E, Janmey P A, Kwiatkowski D J, Stossel T P, Banno Y, Nozawa Y, Mullauer L, Ishizaki A, Kuzumaki N. Eur J Biochem. 1995;229:615–620. doi: 10.1111/j.1432-1033.1995.tb20505.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson K P, Black J F, Thomson J A, Kim E E, Griffith J P, Murcko M A, Chambers S P, Aldape R A, Raybuck S A, Livingston D J. Nature (London) 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 26.Walker N P C, Talanian R V, Brady K D, Dang L C, Bump N J, Ferenz C R, Franklin S, Ghayur T, Hackett M, Hammill L D, et al. Cell. 1994;78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 27.Rotonda J, Nicholson D W, Fazil K M, Gallant M, Gareau Y, Labelle M, Peterson E P, Rasper D M, Ruel R, Vaillancourt J P, Thornberry N A, Becker J W. Nat Struct Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 28.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–450. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 29.Xue D, Shaham S, Horvitz H R. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- 30.Kuida K, Zheng T S, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell R A. Nature (London) 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa J-I, Kamada S, Kamiike W, Shimizu S, Imazu T, Matsuda H, Tsujimoto Y. Cancer Res. 1996;56:1713–1718. [PubMed] [Google Scholar]
- 32.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 34.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 35.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 36.Witke W, Sharpe A H, Hartwig J H, Azuma T, Stossel T P, Kwiatkowski D J. Cell. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- 37.Cheng E H-Y, Kirsch D G, Clem R J, Ravi R, Kastan M B, Bedi A, Ueno K, Hardwick J M. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 38.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli K J, Debatin K-M, Krammer P H, Peter M E. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 40.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 41.Rudel T, Bokoch G M. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 42.Brenner B, Koppenhoefer U, Weinstock C, Linderkamp O, Lang F, Gulbins E. J Biol Chem. 1997;272:22173–22181. doi: 10.1074/jbc.272.35.22173. [DOI] [PubMed] [Google Scholar]
- 43.Juo P, Kuo C, Reynolds S E, Konz R F, Raingeaud J, Davis R J, Biemann H-P, Blenis J. Mol Cell Biol. 1997;17:24–35. doi: 10.1128/mcb.17.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenczowski J M, Dominguez L, Eder A M, King L B, Zacharchuk C M, Ashwell J D. Mol Cell Biol. 1997;17:170–181. doi: 10.1128/mcb.17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang S, Jiang Y, Li Z, Nishida E, Mathias P, Lin S, Ulevitch R J, Nemerow G R, Han J. Immunity. 1997;6:739–749. doi: 10.1016/s1074-7613(00)80449-5. [DOI] [PubMed] [Google Scholar]
- 47.Enari M, Hug H, Nagata S. Nature (London) 1995;375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- 48.Los M, Van de Craen M, Penning L C, Shenk H, Westendorp M, Baeuerle P A, Droge W, Krammer P H, Fiers W, Schulze-Osthoff K. Nature (London) 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- 49.Schlegel J, Peters I, Orrenius S, Miller D K, Thornberry N A, Yamin T-T, Nicholson D W. J Biol Chem. 1996;271:1841–1844. doi: 10.1074/jbc.271.4.1841. [DOI] [PubMed] [Google Scholar]
- 50.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 51.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 53.Xue D, Horvitz H R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 54.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]