Abstract
2-Hydroxyadenine (2-OH-A), a product of DNA oxidation, is a potential source of mutations. We investigated how representative DNA polymerases from the A, B and Y families dealt with 2-OH-A in primer extension experiments. A template 2-OH-A reduced the rate of incorporation by DNA polymerase α (Pol α) and Klenow fragment (Kfexo−). Two Y family DNA polymerases, human polymerase η (Pol η) and the archeal Dpo4 polymerase were affected differently. Bypass by Pol η was very inefficient whereas Dpo4 efficiently replicated 2-OH-A. Replication of a template 2-OH-A by both enzymes was mutagenic and caused base substitutions. Dpo4 additionally introduced single base deletions. Thermodynamic analysis showed that 2-OH-A forms stable base pairs with T, C and G, and to a lesser extent with A. Oligonucleotides containing 2-OH-A base pairs, including the preferred 2-OH-A:T, were recognized by the human MutSα mismatch repair (MMR). MutSα also recognized 2-OH-A located in a repeat sequence that mimics a frameshift intermediate.
Keywords: 2-Hydroxyadenine, Mismatch repair, Replication, Y family polymerases
1. Introduction
Reactive oxygen species (ROS) can cause oxidative damage to DNA, producing chemical changes to pyrimidine and purine bases, abasic sites and single and double strand breaks. In addition, ROS can react with the DNA precursors in the dNTP pool providing another source of oxidative DNA damage. The relative numbers of different lesions, their intrinsic miscoding potential and the efficacy of dedicated repair systems are all factors that control the final mutational load and the development of oxidation stress-related diseases. Among oxidized DNA bases, 8-oxo-7,8-dihydroguanine (8-oxoG) has attracted major attention because of its potent miscoding ability. In vitro studies have shown that template 8-oxoG can direct incorporation by DNA polymerases of either C or A [1]. The latter results in GC>TA transversions [2]. In addition, DNA polymerases can utilise 8-oxodGTP and the resulting incorporation of 8-oxoG opposite A causes AT>CG transversions and contributes to the general oxidative and mutagenic load [3,4].
In addition to 8-oxoG, sources of oxidation such as Fenton-type reagents, γ-rays or antitumor drugs [5–7] all increase the levels of 2-hydroxyadenine (2-OH-A) in DNA. 2-OH-A is also potentially miscoding [8,9]. Replication in bacteria or mammalian cells of shuttle vectors containing a single 2-OH-A produces a broader spectrum of mutations than that produced by DNA 8-oxoG [9,10]. 2-OH-A has received less attention, probably because steady-state levels of 2-OH-A residues in cellular DNA are estimated to be more than one order of magnitude lower than those of 8-oxoG (1/107 normal nucleotides) [5,6]. Significantly, the main source of DNA 2-OH-A appears to be utilization of 2-OH-dATP during replication whereas in situ oxidation of DNA adenine makes a relatively minor contribution [5]. While no information is available on the excision repair of DNA 2-OH-A, replication-related processing of 2-OH-A, including its incorporation from the oxidized dNTP pool, has been more extensively investigated. Indeed, the major replicative enzyme DNA polymerase α (Pol α) incorporated 2-OH-dATP opposite T and C in the DNA template, while Escherichia coli DNA polymerase I Klenow fragment (Kfexo−) incorporated 2-OH-dATP only opposite T [5]. Thus, 2-OH-dATP might have a mutagenic potential for replicative DNA polymerases. It is well established that 2-OH-dATP is a substrate for hydrolysis by the human MutT homolog, hMTH1. This prevents the incorporation of 2-OH-A into DNA [11]. In addition, MYH, the MutY homolog which excises A incorporated opposite DNA 8-oxoG, also removes 2-OH-A from 2-OH-A:G base pairs. This is consistent with a role for MYH in reversing oxidation-related mismatches generated by incorporation of 2-OH-A [12].
Our previous work demonstrated that the post-replicative mismatch repair (MMR) pathway also helps to regulate the steady-state level of DNA 8-oxoG by removing the oxidized base from the nascent DNA strand [13,14]. Furthermore, over-expression of hMTH1 in MMR-defective mouse and human cells reduces the level of DNA 8-oxoG and significantly attenuates their characteristic mutator phenotype [15]. Mutation and microsatellite instability analysis indicated that a significant fraction of the oxidation-related mutations that were subject to correction by MMR occurred at A:T base pairs [15]. In particular, AT>TA, AT>GC mutations and frameshifts in runs of As were all affected. Since hMTH1 acts on both 2-OH-dATP and 8-oxodGTP [16], its expression could influence mutation by either of the oxidized purines, suggesting that DNA 2-OH-A might make a significant contribution to the mutational burden.
As a first step to clarifying the possible involvement of DNA 2-OH-A in oxidation-related mutagenesis, we have investigated some of its biochemical and physical properties. We examined the effect of 2-OH-A on replication in vitro by replicative and translesion synthesis (TLS) DNA polymerases. 2-OH-A miscoding was investigated in two unrelated DNA sequences, in A repeats or in random sequences, in which its effect on the thermal stability of DNA duplexes was quantified. The ability of a purified human MutSα DNA mismatch binding complex to recognize 2-OH-A-containing base pairs was compared in random sequences and in repetitive DNA sequences that represent frameshift intermediates. Our findings indicate that 2-OH-A is a block for replicative DNA polymerases and its bypass by TLS polymerases is mutagenic. In addition the evidence that MutSα can recognize 2-OH-A-containing base pairs, including frameshift intermediates, suggest that MMR might help to counteract the effects of 2-OH-A incorporated from the oxidized pool.
2. Materials and methods
2.1. Oligonucleotide synthesis
Oligonucleotides were synthesized by MWG-Biotech AG. 6-Carboxyfluorescein (6-FAM) labelled and 2-OH-A containing oligonucleotides were synthesized by the Eurogentec S.A. All oligonucleotides were further purified by denaturing polyacrylamide gels (PAGE).
2.2. Primer extension reactions
In standard primer extension experiments, 6-FAM labelled primers were annealed to the template strands in a 1:1 molar ratio. 6-FAM labelled DNA duplexes (50 nM) were initially pre-incubated with 0.2 pmol of mammalian Pol α in 25mM Tris–HCl (pH 8.0), 0.5mM DTT, 0.25 mg/ml BSA, 10mM MgCl2 buffer for 1min. Nucleotides were then added as specified in the figure legends and the reaction continued for 10 min at 37 °C. Reactions were stopped by addition of gel loading buffer (USB Corporation) (95% formamide, 20mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), the products were denatured at 95 °C for 5min and separated on denaturing 20% PAGE. Pol α was purified from HeLa cells as described in Ref. [17]. For dNTPs incorporation and extension by Kfexo− (New England Biolabs) 6-FAM labelled DNA substrates (50 nM) were incubated with the enzyme (2.5 nM) and 30μM of each triphosphate at 37 °C in a buffer containing 20mM Tris–HCl (pH 7.7), 2mM MgCl2, 2mM DTT. After 0.5–5 min, an equimolar dNTP mixture was added and incubated for 5min. Primer/template (50 nM) were pre-incubated with 150nM DNA polymerase 4 (Dpo4), purified as described in Gruz et al. [18], at 55 °C in a buffer containing 30mM potassium phosphate, pH 7.4, 7.5mM MgCl2, 1.25mM β-mercaptoethanol and 5% glycerol. After 3min the dNTP (50μM) were added and incubated at 55 °C for 15 min. The subsequent elongation was performed by adding 50μM of dNTPs for further 15 min. Fluorescent bands were visualized by Typhoon 9200 Gel Imager (Amersham Bio-sciences Europe GmbH) and quantitated by ImageQuant TL software.
2.3. Kinetic analysis
Experiments with Pol α, Kfexo− and Dpo4 were performed under the conditions described above using 0.01–100μM dNTP, 0.01–300μM dNTP and 1–300μM dNTP, respectively. Data points were derived from the analysis of the intensities of the products bands. The values of integrated gel band intensities in dependence of the nucleotide substrate concentrations ([dNTP]) were fitted to the equation:
where T is the target site, the template position of interest; = the sum of the integrated intensities at positions T, T + 1· · ·T + n.
Before being inserted in the above equation, the intensities of the single bands of interest were first normalized by dividing for the total intensity of the lane. This reduced the variability due to manual gel loading. An empty portion of the gel was scanned and the resulting value was subtracted as background. The goodness of fit of the interpolated curve was assessed by computer-aided calculation of the sum of squares of errors SSE and the correlation coefficient R2. Interpolation, SSE, R2 and standard errors determination were done with the computer programs GraphPadPrism and Kaleidagraph.
2.4. Bypass efficiency assay
Reaction conditions for Dpo4 and polymerase η (Pol η) were the same described previously [19]. Reaction conditions for Kfexo− were as recommended by the manufacturer using 25μM dNTPs. A truncated Pol η (residues 1–434) was overexpressed in E. coli using a modified pGEX4T3 vector that codes for a GST-Pol η fusion with a TEV cleavage site between the two proteins. Polymerase was purified by batch binding to glutathione Sepharose 4B, on-resin cleavage with TEV and MonoS column chromatography, with similar conditions to those described previously for the full length protein [20]. Dpo4 was purified as previously described [21]. The DNA substrate was a 45-mer template (5′-CCAGCTCGGTACCGGGTTAGCCTTTGGAGTCGACCTGCAGAAATT; underlined A is site of 2-OH-A) annealed to a 24-mer 32P end-labelled primer (5′-AATTTCTGCAGGTCGACTCCAAAG). Reactions were prepared on ice without enzyme, preheated for 30 s to the reaction temperature, and polymerase added to initiate synthesis. Samples were removed at the indicated times, added to an equal volume of formamide loading buffer and processed as described above for analysis by 12% PAGE and scanning densitometry with a Phospholmager. All reactions contained 4 pmol substrate and the substrate:enzyme ratios: Kfexo− = 1000:1, Dpo4 = 1000:1, Pol η = 750:1. Determination of single hit conditions and calculation of bypass efficiency were as described previously [21].
2.5. Bypass fidelity assay
Lesion bypass fidelity assays were performed with a substrate prepared with an unlabelled primer, using instead an internal 32P-dCTP label, as described previously [21,22]. Reaction conditions were the same as for the bypass efficiency reactions, except that a 5:1 substrate:enzyme ratio was used with a 15min incubation for all enzymes. Processing of the full length synthesis products and determination of plaque colour frequency and corresponding error rates were performed as previously described [21,22].
2.6. UV melting
Absorbance versus temperature changes were measured at 260nm by means of a Cary 3 spectrophotometer equipped with a Peltier device for temperature control. Samples resuspended in Tris–HCl 10 mM pH 7.2, MgCl2 2mM were placed in 1 cm path length quartz cells. The heating rate was 0.5 °C/min and data points were recorded every 0.2 °C. Absorbance values were corrected for the water thermal expansion and normalized at the absorption of 1 OD at 5 °C. Thermodynamic parameters were evaluated by extracting information by single equilibrium transition curves and data analysis was performed according to Breslauer [23].
2.7. MutSα purification and bandshift
MutSα was prepared from approximately 2 × 1010 Raji cells as previously described [24]. The final concentrated Q sepharose pool contained approximately 5 pmol MutSα per milliter. For bandshift assays, one unit of MutSα was defined as 5 fmol. Bandshift experiments were carried out with 32P-end labelled oligonucleotide duplexes as previously described. Briefly, MutSα (2–10 units) was pre-incubated (5 min at 20 °C) with 2 pmol non-radioactive matched competitor duplex in 20μl reaction buffer containing 25mM Hepes KOH pH 8.0, 0.5 mM EDTA, 0.1 mM ZnCl2, 10% glycerol, 50μg poly(dI:dC). The 20 fmol substrate duplex was added and incubation continued for a further 20 min. Products were analysed by PAGE on 6% non-denaturing gels.
3. Results
3.1. Replication of 2-OH-A-containing templates by different DNA polymerases
Since miscoding by a DNA lesion can be influenced by the type of DNA polymerase [25,26] and the sequence context, we examined the ability of A, B and Y family DNA polymerases to bypass a template 2-OH-A in two different sequences. Because we identified oxidation-related mutations at A:T base pairs in microsatellites formed by A-runs, we placed a single 2-OH-A in the middle of an A run within a 36 mer (6A*, repeated sequence). In the second DNA substrate, a single 2-OH-A replaced the A repeat, and the 15 nt flanking sequence on both 5′ and 3′ sides was retained (A*, random sequence). In control oligonucleotides, A replaced 2-OH-A. The results obtained with each enzyme are detailed in the next sections.
3.1.1. Human DNA polymerase α
The ability of the human B family Pol α to replicate 2-OH-A located in the A* and the 6A* repeat was investigated in primer extension experiments, using primers that terminated one base immediately before the lesion (Fig. 1). Following incubation in the presence of a single dNTP, Pol α incorporated exclusively T opposite 2-OH-A in either sequence (Fig. 1A and B) and there was no detectable incorporation of any of the other three bases. Although 2-OH-A retained the coding specificity of undamaged A, its coding efficiency – the ability to instruct the polymerase to incorporate the complementary base – was significantly reduced (Fig. 1, supplementary data). The apparent incorporation efficiency (kcat/Km) for T opposite 2-OH-A was 17.5- and 5.5-fold lower than opposite A in the random and in the repeated sequence, respectively (Fig. 1C). Thus, Pol α preferentially incorporates T opposite 2-OH-A, but with lower efficiency than opposite a normal A in both sequence contexts.
Fig. 1.
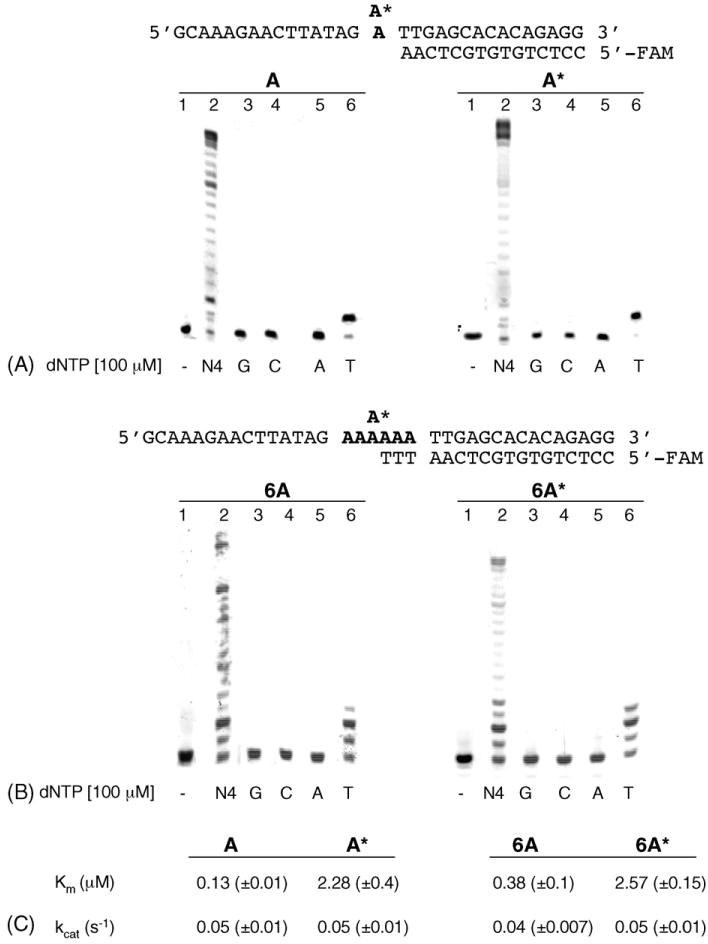
DNA polymerase α: incorporation of dNTP opposite 2-OH-A. (A) Random sequence. Primer/templates (50 nM) were pre-incubated with Pol α. After 1min, reactions were initiated by the addition of a single triphosphate (100μM) and continued for 10 min. Reactions products were analysed by denaturing PAGE as described in Section 2. Controls (lanes 1) were incubated without enzyme or dNTP. A* = 2-OH-A; (B) 6A or 6A* repeat sequence. The experimental conditions were the same used for the random sequence. (C) Kinetic parameters for dTTP incorporation opposite A or 2-OH-A by Pol α in the random and in the repeated sequence. Km and kcat values were evaluated as described in Section 2.
3.1.2. E. coli DNA polymerase I (Kfexo−)
In contrast to Pol α, the Klenow fragment (Kfexo−) of E. coli DNA polymerase I is able to incorporate various bases opposite a template 2-OH-A [8]. Using the same experimental approach employed for Pol α, we compared the ability of Kfexo− to replicate a template 2-OH-A in the random and in the repeated sequence. The kinetic parameters for correct and incorrect nucleotide insertion by Kfexo− are summarized in Table 1A. Primer extension was observed in the presence of each of the four dNTPs but with very different efficiencies. The degree of dNTP selectivity differed for the random and the 6A* sequence. T was preferentially inserted opposite 2-OH-A in both, although the insertion efficiency was, respectively, 70- and 56-fold lower than opposite A. For the other dNTPs, the preferential order of incorporation in the A* random sequence was T≫G=C≫A with utilization of dATP three orders of magnitude lower than TTP. In the 6A* substrate, the discrimination against A was relaxed and both purines were inserted with similar efficiencies and about three-fold better than C. The preferred order was T≫G = A>C (Table 1A).
Table 1.
Kinetic parameters for dNTP incorporation opposite adenine or 2-OH-adenine by Kfexo− (A) and Dpo4 (B) in a random and in a repeated sequence
| A | ||||
|
| ||||
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1 × 105) | Selectivitya |
|
| ||||
| Kfexo− | ||||
| A | ||||
| dTTP | 0.0042(±0.0004) | 0.6(±0.006) | 1428(±14.8) | 1 |
| A* | ||||
| dTTP | 0.21(±0.04) | 0.43(±0.02) | 20.5(±0.49) | 69.7 |
| dATP | 18.1(±3.6) | 0.049(±0.002) | 0.03 | 47600 |
| dCTP | 27.7(±4.5) | 0.19(±0.01) | 0.67 | 2131 |
| dGTP | 2.1(±0.2) | 0.141(±0.003) | 0.7 | 2040 |
| 6A | ||||
| dTTP | 0.06(±0.007) | 1.89(±0.04) | 315(±3.68) | 1 |
| 6A* | ||||
| dTTP | 0.55(±0.11) | 0.31(±0.01) | 5.6(±0.1) | 56.2 |
| dATP | 4.93(±0.97) | 0.04(±0.002) | 0.081(±0.002) | 3888.9 |
| dCTP | 13.9(±1.7) | 0.039(±0.001) | 0.028 | 11250 |
| dGTP | 3.7(±0.7) | 0.036(±0.001) | 0.09(±0.002) | 3500 |
| B | ||||
|
| ||||
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1 × 104) | Selectivitya |
|
| ||||
| Dpo4 | ||||
| A | ||||
| dTTP | 0.79(±0.1) | 0.035 | 4.43 | 1 |
| A* | ||||
| dTTP | 11.3(±1.7) | 0.017(±0.001) | 0.15 | 29.5 |
| dATP | 85.4(±13) | 0.031(±0.003) | 0.03 | 147.7 |
| dCTP | 69.2(±11) | 0.038(±0.005) | 0.05 | 88.6 |
| dGTP | N.D. | |||
| 6A | ||||
| dTTP | 8.8(±1.4) | 0.11(±0.005) | 1.25 | 1 |
| 6A* | ||||
| dTTP | 32.9(±4.5) | 0.073(±0.006) | 0.22 | 5.7 |
| dATP | 41.7(±6.5) | 0.023(±0.002) | 0.05 | 25 |
| dCTP | N.D.b | |||
| dGTP | N.D. | |||
Selectivity is defined as (kcat/Km)TTP/(kcat/Km)dXTP, where X is A, G or C. The value represents the number of correct (TTP) incorporation events for each incorrect (dXTP) incorporation.
Not detectable.
We also examined whether Kfexo− could elongate 2-OH-A-containing base pairs. Two-stage reactions were performed. In the first step, Kfexo− was incubated with primer/templates and a single triphosphate to allow incorporation. To monitor the efficiency of elongation from the resulting 3′-terminal 2-OH-A base pair, a second incubation was carried out in the presence of all four dNTPs (Fig. 2, supplementary data). Kfexo− efficiently elongated all terminal 2-OH-A base pairs in the random sequence. In the 6A* sequence, terminal 2-OH-A:C and 2-OH-A:A pairs impeded extension (Fig. 2, supplementary data). Similar results were obtained in an alternative approach, in which the extension of synthetic primers generating different 3′ terminal 2-OH-A-containing base pairs by Kfexo− and all four dNTPs was assayed (Fig. 2A). In the 6A* sequence Kfexo−efficiently elongated a terminal 2-OH-A:T or 2-OH-A:G pair whereas a 2-OH-A:A and, to a more modest extent a 2-OH-A:C pair, prevented elongation by Kfexo−.
Fig. 2.
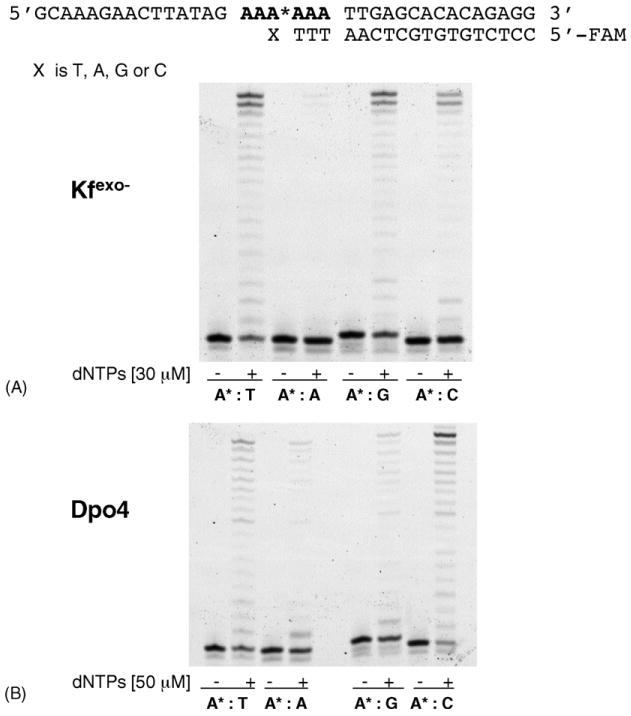
Elongation of terminal 2-OH-A:T, 2-OH-A: A, 2-OH-A:G and 2-OH-A:C base pairs. (A) Elongation of 2-OH-A containing terminal mismatches in the 6A* sequence by Kfexo−. Reactions contained 50nM primer/template and 2.5nM Kfexo− supplemented with an equimolar mixture of dNTPs (30μM). Controls were incubated without enzyme or dNTP. (B) Elongation of 2-OH-A containing terminal mismatches in the 6A* sequence by Dpo4. Primer/template (50 nM) were pre-incubated with 150nM Dpo4 enzyme at 55 °C for 3min. The elongation was performed by adding 50μM of dNTPs for 15 min.
We conclude that replication of 2-OH-A by Kfexo−, as with Pol α, is also relatively error-free, and T is the preferentially incorporated base. Unlike Pol α, Kfexo− exhibits a somewhat more relaxed specificity and other base pairs – notably 2-OH-A: G – are also formed. Both the formation and elongation of these promutagenic base pairs is influenced by sequence context and these parameters differ significantly between the random and repeat sequence.
3.1.3. Sulfolobus solfataricus Dpo polymerase 4
S. solfataricus Dpo4 is an archeal Y family DNA polymerase [27]. With a single dNTP, Dpo4 inserted T, A or C opposite 2-OH-A in the A* random sequence and the preferential order of incorporation was T>C>A (Table 1B). As expected, incorporation by Dpo4 was less selective than Kfexo− and dNTP utilization varied by only 3–5-fold. Surprisingly, however, addition of G was undetectable.
A different result was obtained with the 6A* repeat sequence in which Dpo4 inserted predominantly T or A. Kinetic parameters indicated that T incorporation opposite 2-OH-A is only slightly lower (5.7-fold) than opposite A, while misincorporation of A is further decreased (25-fold) (Table 1B).
In this repeat sequence, Dpo4 efficiently elongated primers creating terminal 2-OH-A:T or 2-OH-A:C pairs, whereas 2-OH-A:G and 2-OH-A:A constituted a significant block (Fig. 2B). In the random sequence, none of the terminal mismatches constituted a significant block to elongation by Dpo4 (Fig. 3, supplementary data).
Fig. 3.
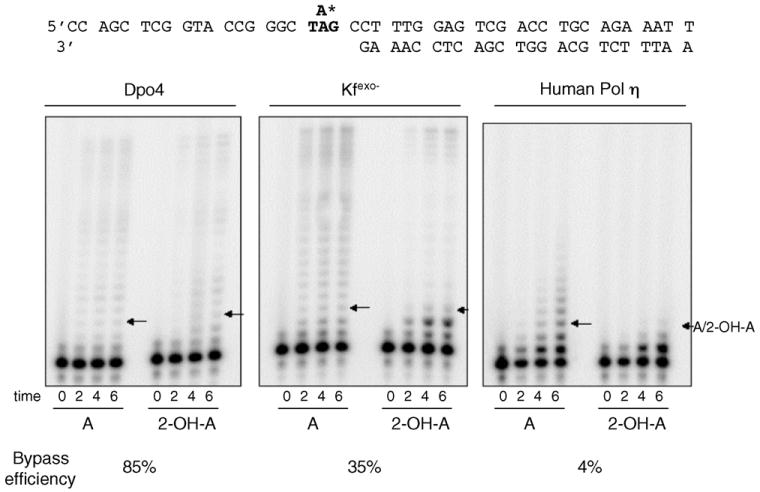
Products of 2-OH-A bypass reactions. Reactions shown are for bypass reactions with Dpo4, Kfexo− and human DNA Pol η (left, middle and right panel, respectively). Each panel shows bypass of an undamaged A and 2-OH-A within a 5′-TAG substrate (as indicated in the sequence). An arrow indicates the location of the lesion.
A template 2-OH-A in a random sequence is therefore quite efficiently bypassed by Dpo4. Bypass occurs with a significantly reduced fidelity via the formation of frequent 2-OH-A:C and 2-OH-A:A mispairs. Once formed, each of these terminal mismatches are easily elongated. Dpo4 is also considerably influenced by the sequence context of the 2-OH-A. In the 6A* repeat sequence, bypass of 2-OH-A by Dpo4 is more efficient. Its inability to extend the frequent terminal 2-OH-A:A mismatches indicates that 2-OH-A bypass in a repeat sequence is also likely to be more error free.
3.2. 2-OH-A differentially affects the bypass efficiency of Dpo4 and human DNA polymerase η
Bypass of 2-OH-A by Y family DNA pols was also investigated by a different approach. We used reaction conditions that reflect a single cycle of DNA synthesis with a primer terminus located such that multiple incorporations are required prior to the lesion being encountered [21]. Short incubation times and large substrate excess are chosen so that each DNA molecule is acted on only once during the time course studied. The advantage to this ‘single hit’ condition is that quantitative measurements of insertion efficiency opposite the lesion, extension efficiency from the damaged primer terminus and bypass efficiency (defined here as incorporation opposite at least one undamaged base beyond the lesion) can be made by comparing synthesis on the damaged and undamaged templates. All reactions described here were confirmed to be under single hit conditions that yield fainter band intensities (Fig. 3) than observed when multiple cycles of synthesis occur (Figs. 1 and supplementary data).
In these assays, bypass efficiency of 2-OH-A by Dpo4 was high, 85% of the efficiency of copying the equivalent undamaged A, and there was no obvious pause at the site of the lesion (Fig. 3, arrows in left panel). The relative insertion efficiency opposite 2-OH-A and the relative extension efficiency from the damaged terminus were 79 and 88%, respectively, indicating that 2-OH-A is only a minor block to normal synthesis.
The same analysis performed with human Pol η, the prototypical Y family polymerase, indicated that bypass of 2-OH-A by this enzyme was only 4% that of undamaged A (Fig. 3, right panel). In addition, insertion opposite the lesion was also very inefficient and was only 22% of the corresponding value for A. Surprisingly, incorporation by Pol η also appeared to be inhibited at the base preceding the 2-OH-A (note the low intensity of the band immediately below the arrow in Fig. 3, right panel), a property not previously reported for this enzyme.
Kfexo− was assayed under the same single hit conditions to provide a comparison (Fig. 3, middle panel). Bypass efficiency was 35% and in this case, insertion opposite the lesion (note the more intense band at the −1 position), rather than subsequent extension was particularly problematic. Both these findings are in agreement with expectations from the kinetic analysis presented in Table 1. Thus, under conditions of a single polymerase/template encounter, a template 2-OH-A lesion has a minimal impact of replication by Dpo4 whereas synthesis by human Pol η is strongly inhibited. Its overall effect on replication by Kfexo− is intermediate and, as expected, the major impact is on insertion opposite the lesion.
3.3. Fidelity of 2-OH-A bypass by Dpo4, DNA polymerase η and Kfexo−
In order to measure the error rates for a complete lesion bypass event in the presence of all four dNTPs, we used templates corresponding to the N-terminal region of the lacZ α-complementation gene of bacteriophage M13mp2, in which 2-OH-A (or undamaged A) is located in a TAG stop codon. Reactions designed to allow complete synthesis (to the end of the template) were carried out on all substrates and the newly synthesized strand was recovered, hybridized to gapped-duplex M13 molecules, and introduced into the appropriate E. coli strain [21]. Errors in synthesis at either the T, A/2-OH-A or G of the stop codon were detected as dark blue plaques and frameshift errors were detected as colourless plaques. The precise change was then identified by sequencing of mutant plaques. The frequencies of the different plaque phenotypes (Table 1, supplementary data) and the corresponding rates of base substitution and insertion/deletion errors at the lesion site are shown in Fig. 4A. Bypass of 2-OH-A by each of the three enzymes induced large increases in base substitutions (5–90 fold) relative to undamaged A, indicating 2-OH-A acts as a miscoding lesion. In addition, replication by Dpo4, but not Pol η or Kfexo−, produced a six-fold increase in frameshifts at 2-OH-A. These were mostly single base deletions at the site of 2-OH-A.
Fig. 4.
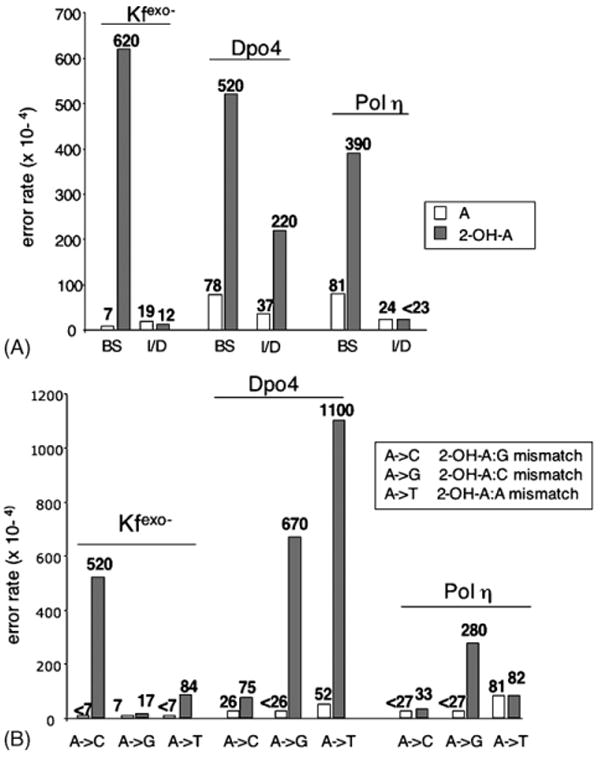
(A) Error rates for undamaged A and 2-OH-A by Dpo4, Kfexo− and human DNA Pol η. The error rates of the different polymerases for undamaged A (open bars) and 2-OH-A (filled bars) are shown. BS, base substitution; I/D, insertion/deletion. (B) Base substitution error rates for Kfexo−, Dpo4 and human DNA Pol η for undamaged A (open bars) and 2-OH-A (filled bars). In the box are indicated the possible 2-OH-A containing mismatches causing the observed mutations.
Each polymerase produced a different spectrum of base substitution mutations at the 2-OH-A site (Fig. 4B). Dpo4 bypass of 2-OH-A was associated with both AT>TA transversions and AT>GC transitions (in an approximately 3:2 ratio). By contrast, bypass by Pol η caused predominantly AT>GC transitions, whereas the majority of changes for Kfexo− were AT>CG transversions, with a smaller contribution from AT>TA.
These findings indicate that replication of 2-OH-A by these three polymerases is mutagenic and that the type of mutation is determined by the polymerase. Furthermore, the mutational spectra are in good general agreement with the incorporation preferences of each polymerase inferred from in vitro experiments (Table 1). Interestingly, Dpo4 was the only polymerase that introduced frameshifts during replication of 2-OH-A in a non-repetitive sequence.
3.4. 2-OH-A effect on DNA secondary structure
DNA base unstacking can underlie failure in base extension during replication [28]. To determine the effects of 2-OH-A on DNA base unstacking as well as on hydrogen bond disruption we carried out thermal stability measurements on DNA duplexes containing the lesion in the random and repeated sequences (Fig. 5).
Fig. 5.
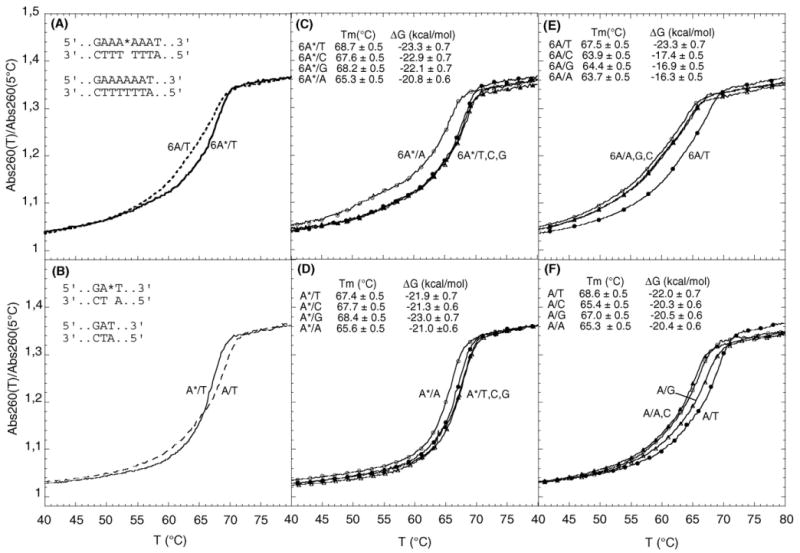
UV denaturation profiles of DNA duplexes with and without 2-OH-A. Effect of 2-OH-A:T pairing in the 6A repeat sequence (panel A) and in a random sequence (panel B); continuous line, 2-OH-A:T; dashed line, A:T. Thermal stability of DNA duplexes with A or 2-OH-A paired with T, G, C or A in the A-repeat sequence (panels E and C, respectively) and in the random sequence (panels F and D). Filled circle, A:T and 2-OH-A:T; empty circle, A:A and 2-OH-A:A; filled triangle, A:C and 2-OH-A:C; empty triangle, A:G and 2-OH-A:G.
In both sequences, an A-containing mismatch produced the expected decrease in Tm (Fig. 5E and F). Replacement of A:T by 2-OH-A:T produced only minor changes in the thermal and thermodynamic stability of either the repeat or random duplexes (Fig. 5A and B). In the random sequence 2-OH-A caused a slight destabilization (Fig. 5B), whereas it had the opposite effect in the repeat sequence (Fig. 5A). 2-OH-A:C or 2-OH-A:G mismatches were without detectable effect and Tm differences in comparison to 2-OH-A:T were within the limits of experimental error (Fig. 5C and D).
The data indicate that 2-OH-dA can form stable base pairs with T and that the sequence context has a role in stacking optimization with stacking favoured in a repetitive A sequence. In contrast to unmodified A, pairing of 2-OH-A with the other bases, with a single exception of A, are relatively favoured. The adoption of a more suitable tautomeric form of the 2-OH-A in base pairs might be responsible for this effect [29].
3.5. Recognition by the hMutSα complex of 2-OH-A containing DNA
Because oxidation-related frameshifts at A repeat sequences are influenced by MMR [15], we examined whether purified MutSα recognizes 2-OH-A-containing base pairs. Duplexes based on the 6A* oligonucleotide in which the 2-OH-A strand was annealed to a complementary strand containing 6, 5 or 7 Ts opposite the A6 sequence were used as substrates for binding by MutSα. These represent frameshift intermediates with an insertion/deletion loop of one base. Band-shift analysis indicated that neither the 2-OH-A:T base pair in the 6A*:6T duplex nor an A:T pair in the control duplex was recognized by MutSα (Fig. 6A). In contrast, we observed significant binding to a duplex containing an extra A in the 2-OH-A-containing strand (6A*:5T). Comparable binding was observed to the duplex in which an extra T was positioned opposite the 2-OH-A-containing strand (6A*:7T) (Fig. 6A). In each case, binding was similar to that observed with duplexes containing either an extra unmodified A or T.
Fig. 6.
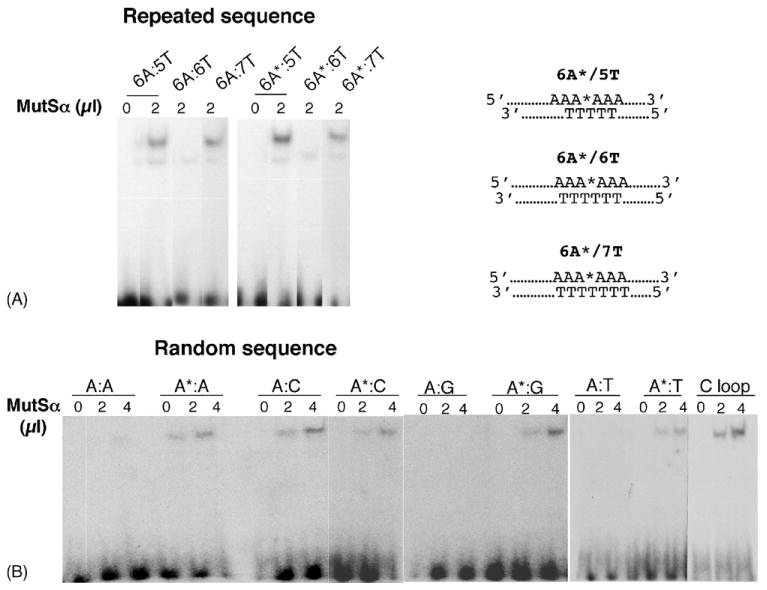
MutSα binding to 2-OH-A. End-labelled oligonucleotide duplexes of the repeated sequences shown in panel A were incubated with MutSα. In panel B is shown MutSα binding to 2-OH-A in random sequences. Products were analysed by non-denaturing PAGE as described in Section 2.
In a second series of experiments, we examined binding to 2-OH-A-containing base pairs in non-repetitive sequences. In this case, 2-OH-A provoked significant recognition by MutSα. Of particular note, binding to a 2-OH-A:T base pair was comparable to that seen with other 2-OH-A-containing mispairs (Fig. 6B). Under the same experimental conditions, there was no detectable binding by MutSα to the control A:T duplex. Recognition of a 2-OH-A:T base pair was also observed with a second series of duplexes based on an unrelated random sequence (data not shown).
These findings indicate that MMR might be engaged at 2-OH-A base pairs. MutSα recognition of 2-OH-A-containing structures resembling slipped-mispaired intermediates suggests a role in counteracting frameshifts caused by these oxidized bases in repetitive sequences. In addition, the ability of MutSα to recognize 2-OH-A containing mispairs is consistent with an involvement in suppressing base substitution mutations caused by oxidatively damaged adenine.
4. Discussion
Our previous analysis of oxidation-related spontaneous mutations in MMR deficient cells identified 2-OH-A as a potentially significant contributor to several classes of mutation [15]. Since these included frameshifts in A runs as well as base substitutions, we investigated some of the structural and biological properties of this oxidized purine in repetitive and non repetitive DNA sequence contexts.
Our findings indicate that incorporation opposite 2-OH-A is difficult for both the A family Kfexo− DNA polymerase and the B family replicative Pol α. This distinguishes 2-OH-A from 8-oxoG, which is easily bypassed by several replicative polymerases via the mutagenic incorporation of an A opposite the lesion [1]. Thus, unlike 8-oxo-G, a template 2-OH-A might cause a permanent or transient replication block thereby provoking recruitment of a TLS polymerase. Among the enzymes we tested, Dpo4 was the least sensitive to 2-OH-A, while human Pol η was inefficient in bypassing the lesion. This raises the possibility that of the human Y family polymerases the Dpo4 homolog, polymerase κ, rather than Pol η, might be better equipped to bypass this oxidized purine. The different efficiencies of 2-OH-A bypass by Dpo4 and Pol η resemble their behaviour with AP sites and may reflect differences in their respective “little finger” subdomains [30,31], probably the critical features that allow lesion bypass [21].
Each polymerase showed a strong preference for insertion of T opposite 2-OH-A although our incorporation data suggested a range of alternative permissible base pairings that were affected by the sequence context of the oxidized base. These findings are consistent with the known ability of DNA 2-OH-A to adopt multiple tautomeric forms that are influenced by temperature, solvent polarity and neighbouring bases [32–34]. In particular, a shift from the prevailing keto tautomer (N1-H) towards the enol form (O2H) is likely to affect the probability that the lesion is accommodated as 2-OH-A:T or with other partners through classical W–C bonding or wobble base pairs. They are also consistent with our Tm measurements, which indicated that 2-OH-A forms stable base pairs not only with T, but also with C or G. Thus, the different abilities of DNA polymerases to accommodate otherwise unfavourable 2-OH-A tautomers within their active sites might influence replication fidelity [35].
A previous investigation of mutational spectra in MMR-defective cells indicated that AT>GC and AT>TA base substitutions (and to a minor extent AT>CG) might derive from miscoding by an oxidized purine [15]. These were the most frequent mutations observed following replication of 2-OH-A by Dpo4 and Pol η under conditions of limited polymerase engagement. The same analysis also revealed that frameshifts, with deletion of the 2-OH-A adduct, were significantly increased following replication by Dpo4. This polymerase has a very low frameshift fidelity, which is consistent with its ability to accommodate unpaired nucleotides in the active site [30]. It is also known to generate deletions at noniterated nucleotides [36]. Together with the apparent difficulties experienced by replicative polymerases at a template 2-OH-A, these findings strengthen the likelihood that Y polymerase-mediated TLS occurs at this lesion.
Little or no information is available on the repair of 2-OH-A formed by in situ oxidation of DNA adenine. We show here that the major human MMR recognition complex, MutSα, recognizes 2-OH-A-containing oligonucleotides. Recognition by MutSα was context dependent. MutSα efficiently recognized oligonucleotide duplexes containing 2-OH-A in structures mimicking insertion/deletion loop (IDL) within a 6A repeat. In this regard, the behaviour of the oxidized adenine was indistinguishable from its normal homolog. Within the same type of IDL structure, the behaviour of 2-OH-A differs from DNA 8-oxoG, which appears to assume a conformation that renders it invisible to MutSα [37]. The ability of MutSα to recognize 2-OH-A within an IDL context may have relevance in vivo as it is consistent with the apparent contribution of the oxidized base to microsatellite instability at the A26 BAT26 sequence in MMR-defective cells [15].
Although duplexes containing 8-oxoG:C base pairs are not recognized [38,39], MutSα bound each of the four 2-OH-A-containing base pairs to a similar extent. Alterations in base geometry and local flexibility are likely to be major determinants of mismatch recognition [28,40]. Recognition of 2-OH-A:T pairs might again reflect the unique ability of 2-OH-A to adopt multiple tautomeric forms, some of which are associated with wobble base pairs and local distortion [33]. A 2-OH-A:T pair in a fully paired A6 repeat sequence was not recognized by MutSα, however. An repeats of this kind are known to assume unusual conformations in which the A:T base pairs show large propeller twist angles and form bifurcated hydrogen bonds involving the N6 amino group of A and the O4 atoms of two adjacent Ts in the opposite strand. We speculate that this particular structural arrangement might confer a greater resistance to deformation and thereby prevent DNA from adopting the structural changes needed to trigger MutSα recognition.
Notwithstanding the precise mechanism by which MutSα recognizes 2-OH-A-containing base pairs, our findings clearly indicate how MMR might help control the steady-state levels of DNA 2-OH-A. The major source of DNA 2-OH-A is acknowledged to be the oxidized dNTP pool [5]. MMR would reverse this incorporation and prevent the accumulation of 2-OH-A in DNA. This role is consistent with the higher levels of DNA 2-OH-A observed in msh2−/− ES cells in comparison to wild type ES cells [41].
In summary, we have shown that replication fork block is the likely outcome of a replicative DNA polymerase encountering a template 2-OH-A. Our findings indicate that a specialized bypass polymerase might overcome the block at the expense of replication fidelity. In addition, MutSα recognition of 2-OH-A-containing substrates indicates that MMR might contribute to mutation avoidance by acting on 2-OH-A-containing base pairs.
Acknowledgments
This work has been partially supported by grants from AIRC, ISS/NIH, FIRB to MB, by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to TK, from a grant-in-aid for international collaborative research SH34407 (Japan Health Science Foundation) to MY and MB, by the CARIPLO Foundation project “Oncogenetica e Proteomica della Replicazione” (2003.1663/10.8441) to GM. We would like to thank Dr. Lars Pedersen of the NIEHS for construction and purification of Pol η.
Abbreviations
- ROS
Reactive oxygen species
- 8-oxoG
8-oxo-7,8-dihydroguanine
- 2-OH-A
2-hydroxyadenine
- MMR
Mismatch repair
- 6-FAM
6-carboxyfluorescein
- Pol α
Polymerase α
- Kfexo−
Klenow fragment
- Dpo4
DNA polymerase 4
- Pol η
Polymerase η
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.dnarep.2006.11.002.
References
- 1.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 2.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G>T and A>C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 3.Pavlov YI, Minnick DT, Izuta S, Kunkel TA. DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry. 1994;33:4695–4701. doi: 10.1021/bi00181a029. [DOI] [PubMed] [Google Scholar]
- 4.Inoue M, Kamiya H, Fujikawa K, Ootsuyama Y, Murata-Kamiya N, Osaki T, Yasumoto K, Kasai H. Induction of chromosomal gene mutations in Escherichia coli by direct incorporation of oxidatively damaged nucleotides. New evaluation method for mutagenesis by damaged DNA precursors in vivo. J Biol Chem. 1998;273:11069–11074. doi: 10.1074/jbc.273.18.11069. [DOI] [PubMed] [Google Scholar]
- 5.Kamiya H, Kasai H. Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation. J Biol Chem. 1995;270:19446–19450. doi: 10.1074/jbc.270.33.19446. [DOI] [PubMed] [Google Scholar]
- 6.Frelon S, Douki T, Cadet J. Radical oxidation of the adenine moiety of nucleoside and DNA: 2-hydroxy-2′-deoxyadenosine is a minor decomposition product. Free Radic Res. 2002;36:499–508. doi: 10.1080/10715760290025889. [DOI] [PubMed] [Google Scholar]
- 7.Birincioglu M, Jaruga P, Chowdhury G, Rodriguez H, Dizdaroglu M, Gates KS. DNA base damage by the antitumor agent 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine) J Am Chem Soc. 2003;125:11607–11615. doi: 10.1021/ja0352146. [DOI] [PubMed] [Google Scholar]
- 8.Kamiya H, Ueda T, Ohgi T, Matsukage A, Kasai H. Misincorporation of dAMP opposite 2-hydroxyadenine, an oxidative form of adenine. Nucleic Acids Res. 1995;23:761–766. doi: 10.1093/nar/23.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamiya H, Kasai H. Mutations induced by 2-hydroxyadenine on a shuttle vector during leading and lagging strand synthesis in mammalian cells. Biochemistry. 1997;36:11125–11130. doi: 10.1021/bi970871u. [DOI] [PubMed] [Google Scholar]
- 10.Kamiya H, Kasai H. Substitution and deletion mutations induced by 2-hydroxyadenine in Escherichia coli: effects of sequence contexts in leading and lagging strands. Nucleic Acids Res. 1997;25:304–311. doi: 10.1093/nar/25.2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J Biol Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 12.Ushijima Y, Tominaga Y, Miura T, Tsuchimoto D, Sakumi K, Nakabeppu Y. A functional analysis of the DNA glycosylase activity of mouse MUTYH protein excising 2-hydroxyadenine opposite guanine in DNA. Nucleic Acids Res. 2005;33:672–682. doi: 10.1093/nar/gki214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colussi C, Parlanti E, Degan P, Aquilina G, Barnes D, Macpherson P, Karran P, Crescenzi M, Dogliotti E, Bignami M. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr Biol. 2002;12:912–918. doi: 10.1016/s0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- 14.Russo MT, De Luca G, Degan P, Bignami M. Different DNA repair strategies to combat the threat from 8-oxoguanine. Mutat Res. 2006;10 doi: 10.1016/j.mrfmmm.2006.03.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Russo MT, Blasi MF, Chiera F, Fortini P, Degan P, Macpherson P, Furuichi M, Nakabeppu Y, Karran P, Aquilina G, Bignami M. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol Cell Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 17.Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hubscher U. Biochemical and functional comparison of DNA polymerases alfa, delta and epsilon from calf thymus. J Biol Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 18.Gruz P, Pisani FM, Shimizu M, Yamada M, Hayashi I, Morikawa K, Nohmi T. Synthetic activity of Sso DNA polymerase Y1, an archaeal DinB-like DNA polymerase, is stimulated by processivity factors proliferating cell nuclear antigen and replication factor C. J Biol Chem. 2001;276:47394–47401. doi: 10.1074/jbc.M107213200. [DOI] [PubMed] [Google Scholar]
- 19.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 21.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 22.McCulloch SD, Kunkel TA. Measuring the fidelity of translesion DNA synthesis. Methods Enzymol. 2006;408:341–355. doi: 10.1016/S0076-6879(06)08021-9. [DOI] [PubMed] [Google Scholar]
- 23.Breslauer KJ. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. In: Agraval S, editor. Methods in Molecular Biology. Vol. 26. Humana Press Inc.; Totowa, NJ: 1994. pp. 347–372. [DOI] [PubMed] [Google Scholar]
- 24.Macpherson P, Humbert O, Karran P. Frameshift mismatch recognition by the human MutS alpha complex. Mutat Res. 1998;408:55–66. doi: 10.1016/s0921-8777(98)00017-2. [DOI] [PubMed] [Google Scholar]
- 25.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 27.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 28.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair. 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Kawakami J, Kamiya H, Yasuda K, Fujiki H, Kasai H, Sugimoto N. Thermodynamic stability of base pairs between 2-hydroxyadenine and incoming nucleotides as a determinant of nucleotide incorporation specificity during replication. Nucleic Acids Res. 2001;29:3289–3296. doi: 10.1093/nar/29.16.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 31.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 32.Sepiol J, Kazimierczuk Z, Shugar D. Tautomerism of isoguanosine and solvent-induced keto-enol equilibrium. Z Naturforsch. 1976;[C] 31:361–370. doi: 10.1515/znc-1976-7-803. [DOI] [PubMed] [Google Scholar]
- 33.Robinson H, Gao YG, Bauer C, Roberts C, Switzer C, Wang AH. 2′-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis. Biochemistry. 1998;37:10897–10905. doi: 10.1021/bi980818l. [DOI] [PubMed] [Google Scholar]
- 34.Maciejewska AM, Lichota KD, Kusmierek JT. Neighbouring bases in template influence base-pairing of isoguanine. Biochem J. 2003;369:611–618. doi: 10.1042/BJ20020922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bias JR, Luque FJ, Orozco M. Unique tautomeric properties of isoguanine. J Am Chem Soc. 2004;126:154–164. doi: 10.1021/ja036806r. [DOI] [PubMed] [Google Scholar]
- 36.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J Biol Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 37.Macpherson P, Barone F, Maga G, Mazzei F, Karran P, Bignami M. 8-oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSαlpha. Nucleic Acids Res. 2005;33:5094–5105. doi: 10.1093/nar/gki813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazurek A, Berardini M, Fishel R. Activation of human MutS homologs by 8-oxoguanine DNA damage. J Biol Chem. 2001 doi: 10.1074/jbc.M111269200. Epub. [DOI] [PubMed] [Google Scholar]
- 39.Larson ED, lams K, Drummond JT. Strand-specific processing of 8-oxoguanine by the human mismatch repair pathway: inefficient removal of 8-oxoguanine paired with adenine or cytosine. DNA Repair (Amst) 2003;2:1199–1210. doi: 10.1016/s1568-7864(03)00140-x. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 41.DeWeese TL, Shipman JM, Larrier NA, Buckley NM, Kidd LR, Groopman JD, Cutler RG, te Riele H, Nelson WG. Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc Natl Acad Sci USA. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]


