Abstract
Specific local brain responses, influenced by parasite sequestration and host immune system activation, have been implicated in the development of cerebral malaria. This study assessed whole-brain transcriptional responses over the course of experimental cerebral malaria by comparing genetically resistant and susceptible inbred mouse strains infected with Plasmodium berghei ANKA. Computational methods were used to identify differential patterns of gene expression. Overall, genes that showed the most transcriptional activity were differentially expressed in susceptible mice 1 to 2 days before the onset of characteristic symptoms of cerebral malaria. Most of the differentially expressed genes identified were associated with immune-related gene ontology categories. Further analysis to identify interaction networks and to examine patterns of transcriptional regulation within the set of identified genes implicated a central role for both interferon-regulated processes and apoptosis in the pathogenesis of cerebral malaria. Biological relevance of these genes and pathways was confirmed using quantitative RT-PCR and histopathological examination of the brain for apoptosis. The application of computational biology tools to examine systematically the disease progression in cerebral malaria can identify important transcriptional programs activated during its pathogenesis and may serve as a methodological approach to identify novel targets for therapeutic intervention.
Cerebral complications are a major cause of death associated with Plasmodium falciparum malaria.1 Although a large body of research has sought to further define cerebral malaria (CM) clinically and mechanistically, there are currently no approved adjunctive therapies, and the case-fatality rate remains high despite antimalarial therapy. Evidence supports several hypotheses regarding the pathogenesis of CM. One theory is that the host immune response, which is necessary for parasite clearance, may be too robust in cases of severe and cerebral malaria, causing deleterious effects to the host itself.2 Support for this hypothesis is derived from multiple studies that have associated excessive host pro-inflammatory cytokine production and an imbalance of anti-inflammatory responses with malarial disease severity.3 An alternate hypothesis emphasizes the role of parasite sequestration and localized responses in the brain, rather than systemic inflammation per se, in the pathogenesis of CM.4 Examples of these localized responses in the brain include endothelial dysregulation and cell adhesion molecule up-regulation,5 localized leukocyte and parasitized erythrocyte sequestration,6 vessel occlusion, parasitized erythrocyte rupture (with concomitant release of parasite-derived glycophosphatidylinositol anchors, hemozoin, and other products), microhemorrhages, and blood-brain barrier breakdown.7
Because of the difficulties in predicting the development of CM in malaria patients and in sampling brain tissue, most studies investigating CM pathophysiology have used either autopsy specimens or animal models. Although autopsy studies have examined brain pathology during terminal stages of CM, the use of clinically relevant murine models provides an opportunity to examine CM progression within different and controllable genetic backgrounds. Experimental infection of mice with Plasmodium berghei ANKA (PbA) is a well-established and -characterized model system that replicates several key features of human CM.8,9
The identification of transcriptional responses associated with susceptibility or resistance to CM in the PbA model may provide insight into disease pathogenesis10,11,12,13 and suggest novel intervention strategies to improve outcome. The aim of this study was to use expression profiling to define transcriptional patterns and regulatory pathways that distinguish the host brain response to experimental cerebral malaria in genetically susceptible (C57BL/6) and resistant (BALB/c) mice over acute infection. The results demonstrate that resistant mice have a dampened global transcriptional response compared with susceptible animals. Functional and network analyses revealed interrelated biological network hubs that implicated prominent mechanistic roles for apoptosis and interferon-regulated gene expression in the pathogenesis of CM. Furthermore, this investigative approach, using computational biology methodology, identified potential targets for innovative therapeutic strategies to modulate host response and improve clinical outcome in cerebral malaria.
Materials and Methods
Experimental Design
Animal use protocols were reviewed and approved by the Faculty of Medicine Advisory Committee on Animal Services at the University of Toronto, and all experiments were conducted according to the animal ethics guidelines of the University of Toronto. Male C57BL/6 and BALB/c mice, 8–12 weeks of age, were obtained from Charles River Laboratories (Senneville, QC, Canada). Cryopreserved P. berghei ANKA (MR4, Manassas, VA) was thawed and passaged through naïve C57BL/6 donor mice until parasitemia in the passage animals reached approximately 10%. On day 0, mice were infected by intraperitoneal injection with 5 × 105 freshly isolated PbA parasitized erythrocytes. Parasitemia was monitored daily after day 3 using thin blood smears. Brains were removed for microarray analysis from four mice per strain at day 0 (before infection), and at 1, 3, and 6 days after infection with PbA (n = 8 animals per time point), for a total of 32 mice. Freshly isolated tissue was immediately stored in RNAlater (Ambion/Applied Biosciences, Streetsville, ON, Canada) at −80°C until use.
RNA Isolation
Brains were homogenized in TRIzol Reagent (Invitrogen, Burlington, ON, Canada), and total RNA was isolated according to the manufacturer’s instructions. RNA was further purified to remove genomic DNA contamination and concentrated using an RNeasyPlus Mini kit (Qiagen, Mississauga, ON, Canada). RNA quality was assessed by determining the 26S/18S ratio using a Bioanalyzer 2100 (Agilent, Santa Clara, CA).
Microarray Data Analysis
Details of the experimental design, hybridization protocols, and raw data sets have been deposited in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/projects/geo/; GSE7814) in accordance with Minimum Information About a Microarray Experiment guidelines. Briefly, biotin-labeled cRNA was prepared from total RNA and hybridized to a Mouse Genome 430A 2.0 oligonucleotide microarray (Affymetrix, Santa Clara, CA) according to Affymetrix-recommended protocols at the University Health Network microarray center. The MOE430A 2.0 GeneChip is a mouse whole-genome expression array consisting of more than 22,600 probe sets and including more than 14,000 well-characterized genes. Each of the 32 samples was hybridized on two microarrays, resulting in a total of 64 microarray experiments. Therefore, both biological (n = 4) and technical (n = 2) replicates were performed for CM-susceptible (C57BL/6) and -resistant (BALB/c) mice at each time point. Microarrays were scanned using Affymetrix GeneChip Scanner, and GeneChip Operating Software was used for image analysis. Background adjustment and quantile normalization across all microarrays was performed using the robust multichip average algorithm (RMAExpress).14 After normalization, intensity values for the technical replicates were averaged for each animal, and all subsequent analyses were limited to the resultant 32 composite microarray experiments.
Statistically significant differential gene expression was determined using a recently developed algorithm for time course microarray studies, extraction and analysis of differential gene expression.15,16 The multiple comparisons problem was addressed using the optimal discovery procedure (Q-value), an improvement on false discovery rate analysis.15,16 A given Q-value provides the maximum allowable false discovery rate, and a cut-off value of ≤1% was selected to designate changes in gene expression as significant. Differentially expressed genes during the course of PbA infection were identified between CM-susceptible and -resistant mice. Additionally, significant gene expression between CM-susceptible and -resistant mice at each time point was determined for subsequent gene ontology analysis.
Principal Components Analysis
Multidimensional scaling using principal components was performed based on the covariance matrix of normalized gene expression values, using the TM4 software of The Institute for Genomic Research (Rockville, MD).17 Principal components analysis reduces the complexity of high-dimensional data structures by projecting them into a low-dimensional subspace that accounts for the majority of data variance.
Gene Ontology Analysis
Functional annotation of the differentially expressed genes was obtained from the Gene Ontology Consortium database, based on their respective molecular function, biological process, or cellular component.18 Functional categories enriched within genes that were differentially expressed between CM-susceptible and -resistant mice at each time point and throughout PbA infection were determined using the expression analysis systematic explorer algorithm.19 A variant of the one-tailed Fisher exact probability test based on the hypergeometric distribution was used to calculate P values.
Gene Network Interactome
A gene-gene interaction network was created using the software and database of Ingenuity Systems (Redwood City, CA).20 This knowledge base has been manually compiled from >200,000 full-text, peer-reviewed scientific articles encompassing approximately 10,000 human, 8000 mouse, and 5000 rat genes. A molecular network of direct physical, transcriptional, and enzymatic interactions among these mammalian orthologs has been developed and served as the basis for creating smaller networks from the gene expression data. These networks were constructed around genes with the highest connectivity using an iterative algorithm and then merged together to create the final interactome. The interactome was drawn based on the connectivity matrix of its members using Pajek’s visualization software.21
Transcription Factor Analysis
A computationally based transcription factor analysis on differentially expressed genes during PbA infection was performed. Putative, enriched transcription factors (TFs) that regulate the differentially expressed genes were identified using the promoter integration in microarray analysis algorithm as implemented in the Expander software.22,23 Initially, a systematic search for putative TF-binding sites 1000 bp upstream and 200 bp downstream of the transcription start sites of all genes present on the Affymetrix GeneChip was undertaken based on the position weight matrices of approximately 300 known mammalian TFs (TRANSFAC database).24 Next, over-represented TFs among differentially expressed genes relative to the “background” set of all of the genes present on the Affymetrix GeneChip were statistically determined. The problem of multiple comparisons was addressed by selecting only enriched TFs with Bonferroni-corrected P values <0.01.
Quantitative Real-Time RT-PCR
cDNA was synthesized from 2 μg of total RNA using Superscript II reverse transcriptase with oligo(dT)12–18 primers (Invitrogen). Serial dilutions of mouse genomic DNA were used as standards.25 gDNA standards or cDNA were added to the quantitative PCR containing 1× Power Sybr Green Master Mix (Applied Biosystems, Foster City, CA) and 0.5 μmol/L primers in a final volume of 10 μl. Quantitative PCR was performed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Copy numbers were normalized to four mouse housekeeping genes: Gapdh, Hprt, Sdha, and Ywhaz.26 Forward (fwd) and reverse (rvs) primer sequences are as follows: C4b-fwd, 5′-TCCATAGGTCAGACCCGCAACTT-3′; C4b-rvs, 5′-TCTCCCTTGTTGTCACTGGTTTCC-3′; Ccl12-fwd, 5′-CCCAGTCACGTGCTGTTATAATGT-3′; Ccl12-rvs, 5′-TCAGCTTCCGGACGTGAATCTTCT-3′; Cxcl10-fwd, 5′-ATGAGGGCCATAGGGAAGCTTGAA-3′; Cxcl10-rvs, 5′-TGATCTCAACACGTGGGCAGGATA-3′; GzmA-fwd, 5′-AAACCAGGAACCAGATGCCGAGTA-3′; GzmA-rvs, 5′-CAGAGTTTCAGAGGGAGCTGACTT-3′; Icam1-fwd, 5′-TGGCTGAAAGATGAGCTCGAGAGT-3′; Icam1-rvs, 5′-GCTCAGCTCAAACAGCTTCCAGTT-3′; Stat1-fwd, 5′-GAAGAAAACAACCGTGCTCCTT-3′; Stat1-rvs, 5′-TCCCTGAGGAGAGCAACCAT-3′; Irf1-fwd, 5′-CACACATCGATGGCAAGGGATACT-3′; Irf1-rvs, 5′-TGGTTCCTCTTTGCAGCTGAAGTC-3′; Irf7-fwd, 5′-TCAGAAGCAGCTGCACTACACAGA-3′; Irf7-rvs, 5′-TACCTCCCAGTACACCTTGCACTT-3′; Gapdh-fwd, 5′-TCAACAGCAACTCCCACTCTTCCA-3′; Gapdh-rvs, 5′-TTGTCATTGAGAGCAATGCCAGCC-3′; Hprt-fwd, 5′-GGAGTCCTGTTGATGTTGCCAGTA-3′; Hprt-rvs, 5′-GGGACGCAGCAACTGACATTTCTA-3′; Sdha-fwd, 5′-TCACGTCTACCTGCAGTTGCATCA-3′; Sdha-rvs, 5′-TGACATCCACACCAGCGAAGATCA-3′; Ywhaz-fwd, 5′-AGCAGGCAGAGCGATATGATGACA-3′; and Ywhaz-rvs, 5′-TCCCTGCTCAGTGACAGACTTCAT-3′.
Histology and in Situ End Labeling of Fragmented DNA (TUNEL Immunohistochemistry)
Brains were removed for histopathology from four mice per strain at day 0 (before infection) and day 6. Brains were preserved in 4% paraformaldehyde/PBS. All embedding, sectioning, and histochemistry was performed by the Pathology Research Program at the University Health Network (Toronto, ON, Canada). Terminal deoxynucleotidyl transferase-mediated UTP nick-end labeling (TUNEL) was performed as previously described.27 Briefly, 0.5-cm-thick slices of brain were embedded in paraffin and cut into 4-μm sections. Sections were de-waxed, dehydrated, and then permeabilized with 1% pepsin (Sigma, Oakville, ON, Canada) in 0.01 N HCl at pH 2.0. Endogenous peroxides and biotin activity was blocked using 3% hydrogen peroxide and the avidin/biotin blocking kit (Vector Labs, Burlington, ON, Canada). Biotinylated nucleotides were incorporated into any DNA fragments using DNA Polymerase 1 Large (Klenow) Fragment (Promega/Fisher, Nepean, ON, Canada) and then detected using streptavidin-horseradish peroxidase (ID Labs, London, ON, Canada) and Nova Red substrate (Vector Labs). Slides were counterstained using Mayer’s hematoxylin. To quantify apoptosis, TUNEL-positive cells were counted in a blinded fashion from the entire sections of five slides of brain sections isolated from four PbA-infected C57BL/6 and BALB/c mice at day 6 after infection.
Results
Distinct Transcriptional Signatures Characterize CM-Susceptible and -Resistant Mice During PbA Infection
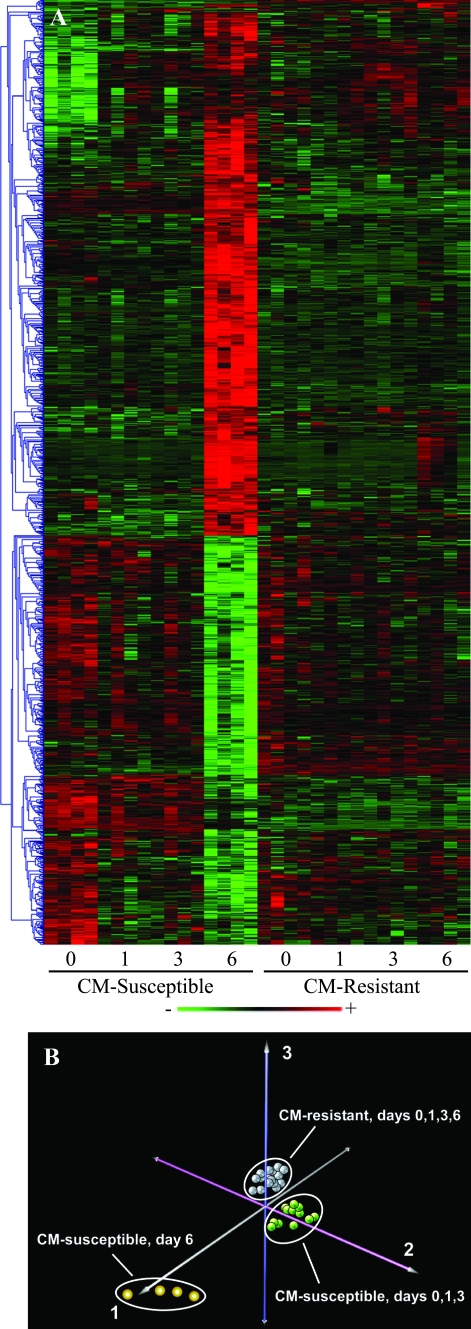
More than 1100 differentially expressed genes were identified in CM-susceptible (C57BL/6) versus -resistant (BALB/c) mice over the course of acute infection at a false discovery rate of ≤1%. A notable feature of the global expression profile was that the majority of these genes underwent intense transcriptional activity late (day 6) during PbA infection in CM-susceptible animals, whereas very few genes changed over time in the CM-resistant mice (Figure 1A). Principal components analysis confirmed these findings by grouping the animals into three clusters based on their gene expression variability (Figure 1B). CM-susceptible mice at day 6 composed the most distinct group followed by CM-susceptible mice (days 0 to 3) and CM-resistant animals (all days). The first principal component, which primarily distinguished CM-susceptible mice on day 6 from the other animals, encompassed more than 50% of the total gene expression variability, implying that the majority of the overall transcriptional variance occurred in this group. No temporal distinction was seen in the expression profiles of CM-resistant mice.
Figure 1.
Overview of differentially expressed genes in CM-susceptible and -resistant mouse brains. Application of the extraction and analysis of differential gene expression time course algorithm to the microarray data set identified more than 1100 genes as differentially expressed between CM-susceptible (C57BL/6) and -resistant (BALB/c) mice. A: Hierarchical clustering of differentially expressed genes revealed that in CM-susceptible animals, the majority underwent intense transcriptional activity late in infection. In the heatmap, red indicates up-regulation and green indicates down-regulation. B: Principal components analysis of the ∼1100 genes identified three prominent clusters of animals: CM-susceptible mice at day 6, CM-susceptible mice (days 0 to 3), and CM-resistant animals (all days). The first principal component, which primarily accounts for CM-susceptible mice on day 6, encompassed more than 50% of the total gene expression variability, reinforcing what was initially observed by clustering the temporal gene-expression profile.
Functional Analysis of Differentially Expressed Genes Reveals Enrichment of Immune, Inflammatory, and Apoptotic Programs
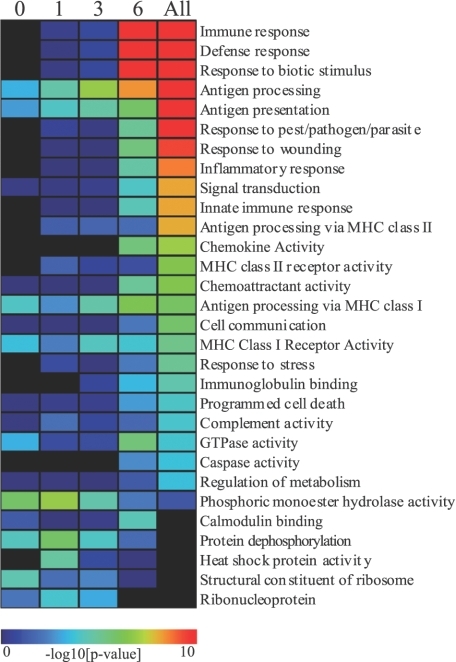
Functional categorization of genes that were differentially expressed between CM-susceptible and -resistant mice at each time point (days 0, 1, 3, and 6) and during the complete time course of infection was performed using expression analysis systematic explorer software. Enrichment of several functional categories became progressively more significant during the course of infection, especially by day 6 (Figure 2). Gene ontology (GO) analysis of differentially expressed genes during the entire course of PbA infection identified various immunological, inflammatory, and pro-apoptotic programs as being highly over-represented (Figure 2, “All” column). These biological modules included innate immune response, antigen processing via major histocompatibility complex classes I and II, chemokine activity, programmed cell death, and caspase activity. Additionally, a limited set of antigen presentation-related GO categories, including antigen presentation, antigen processing, antigen processing via major histocompatibility complex class I, and major histocompatibility complex class I receptor activity, were enriched across the course of infection. Further GO analysis specifically examining gene expression at day 6, when most of the differential transcriptional activity occurred, showed that genes up-regulated in C57BL/6 mice were primarily immune-related, whereas down-regulated genes were associated with developmental processes (Supplemental Table 1, see http://ajp.amjpathol.org).
Figure 2.
Functional categories significantly associated with gene expression differences between C57BL/6 and BALB/c mouse brains. Functional analysis of genes differentially expressed between CM-susceptible and -resistant mice was performed using expression analysis systematic explorer software. Enriched GO categories, which are shown for each time point (columns 1 to 4) and for all differentially expressed genes (column 5), included various immunological, inflammatory, and pro-apoptotic programs. The majority of functional categories were enriched predominantly at day 6; however, some antigen presentation-related GO categories were enriched across infection.
Gene Network Analysis Identifies Hubs of Transcriptional Activity, Including Key Regulators of Interferon Response and Apoptosis
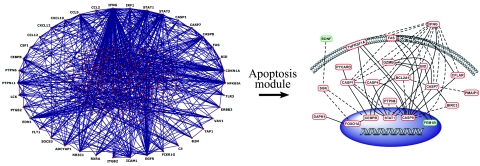
Differentially expressed genes during the time course of PbA infection were linked together based on known gene product interactions using Ingenuity Pathways Analysis software. The resultant gene-gene interaction network or interactome, consisting of 269 genes, was depicted using a visualization tool that shows hubs of high interconnectivity (≥10) on the exterior of the network diagram (Figure 3; Supplemental Table 2, see http://ajp.amjpathol.org). The structure of this interactome was built on several hubs of high interconnectivity, including interferon-γ (Ifn-γ); interferon-responsive factors 1 and 7 (Irf-1 and Ifr-7); signal transducer and activator of transcription 1 and 3 (Stat-1 and Stat-3); caspases 1, 4, 7, and 8; Fas (Cd95); endothelin-1 (Edn-1); and retinoid X receptor-α (Rxra). Recent work suggests that the functional stability of genetic networks is highly dependent on such hubs,28 implying a critical role for these genes in directing the transcriptional response to PbA infection. In fact, 20 of the 38 hubs with connectivity ≥10 have been previously implicated in the pathogenesis of cerebral malaria (Supplemental Table 3, see http://ajp.amjpathol.org).29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47 The majority of apoptosis-related genes within the interactome were differentially up-regulated in the CM-susceptible (C57BL/6) mice relative to CM-resistant (BALB/c) mice (Figure 3), indicating widespread activation of apoptotic pathways in the brains of CM-susceptible mice.
Figure 3.
Interaction network of differentially expressed genes. A gene network based on known gene product interactions was generated using IPA software and displayed using Pajek’s visualization tool (left). Genes that are hubs or nodes of high interconnectivity (≥10) are shown on the outer rim of the network. A complete list of hubs is provided in Supplemental Table 3 (see http://ajp.amjpathol.org). A module within the interactome corresponding to apoptosis-related pathways was amplified and displayed according to subcellular localization of its gene products (right). In the apoptosis module, red ovals are differentially up-regulated and green ovals are differentially down-regulated genes in CM-susceptible (C57BL/6) relative to CM-resistant (BALB/c) mice. The majority of these genes were differentially up-regulated in C57BL/6 mice, demonstrating that there is widespread activation of apoptotic pathways in the brains of CM-susceptible mice.
Promoter Sites for Interferon Response Factors Are Over-Represented among Genes Differentially Expressed during PbA Infection in CM-Susceptible (C57BL/6) Mice
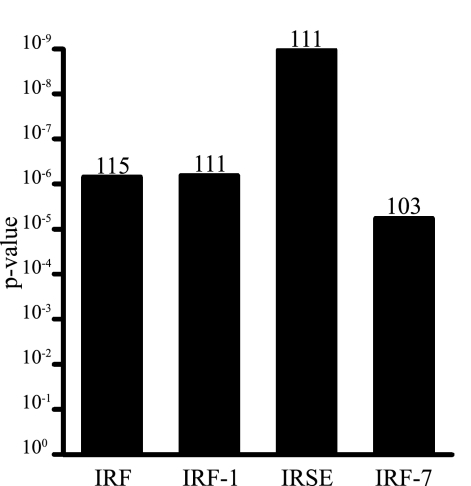
Transcription factor recognition sequences enriched among differentially expressed genes during PbA infection were computationally identified using the promoter integration in microarray analysis algorithm (Figure 4). Four putative promoter sites were highly enriched (Bonferroni corrected P values <0.01), all of which are involved in the regulation of interferon-stimulated gene expression: IRF, IRF-1, IRF-7 and ISRE. These results were consistent with the gene-interaction network analysis that identified Irf-1 and Irf-7 as important hubs for regulating the host response to PbA infection. Another one of the highly enriched promoter sites, interferon-stimulated response element (ISRE), is a key binding site for IRF and STAT families of transcription factors,48,49 including several members of the gene-gene network (Figure 3).
Figure 4.
Transcription factor binding sites over-represented among differentially expressed genes. Analysis of promoter elements in differentially expressed genes using the promoter integration in microarray analysis algorithm identified transcription factors that may regulate transcriptional programs activated in the CM-susceptible versus -resistant brain. The highly enriched promoter sites IRF, IRF-1, IRF-7, and ISRE are all associated with various aspects of interferon-mediated regulation of gene expression. The number of differentially expressed genes recognized by these regulatory factors is show above each column.
Interferon Response Factors and Interferon-Responsive Genes Are Highly Expressed in Brains of CM-Susceptible (C57BL/6) Mice Late in the Course of PbA Infection
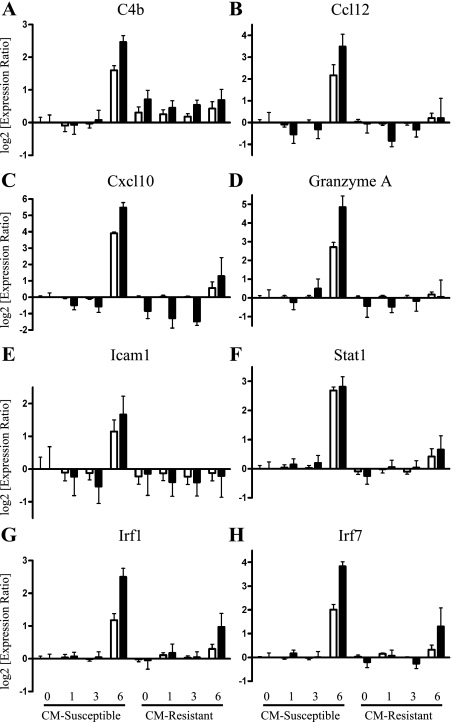
To verify the gene expression data obtained by expression microarray, quantitative real-time RT-PCR was performed on a number of immune-related genes, including complement component 4B (C4b), the chemokines Ccl12 and Cxcl10 (IP10), granzyme A, intercellular adhesion molecule 1 (Icam-1), Irf-1 and Irf-7, and Stat-1 (Figure 5). In general, the transcription profiles of these candidate genes correlated well between expression microarray and PCR analysis. Notably, the majority of these genes are either involved in interferon signaling (eg, Irf-1, Irf-7, and Stat-1) or are interferon inducible (eg, Cxcl10, which contains a ISRE-binding site), and all were identified as being highly up-regulated at day 6 in CM-susceptible C57BL/6 mice compared with all other time points in C57BL/6 and CM-resistant BALB/c mice (Figure 5).
Figure 5.
Real-time RT-PCR validation of expression levels of immune-related transcripts. Several immune-related genes, which were highly up-regulated at day 6 in CM-susceptible compared with resistant mice, were analyzed using quantitative real-time RT-PCR to confirm microarray expression levels. To compare the microarray data (□) with the qRT-PCR data (▪), log2 values of the expression ratio are shown indicating twofold change over baseline. The following genes that were analyzed showed a close correlation between microarray and PCR expression values: complement component 4B (C4b; A), chemokine (C-C motif) ligand 12 (Ccl12; B), chemokine (C-X-C motif) ligand 10 (Cxcl10 or IP10; C), granzyme A (D), intercellular adhesion molecule 1 (Icam1; E), signal transducer and activator of transcription 1 (Stat1; F) interferon regulatory factor 1 (Irf1; G), and interferon regulatory factor 7 (Irf7; H).
Immunohistochemistry Confirms Differential Brain Apoptosis during PbA Infection
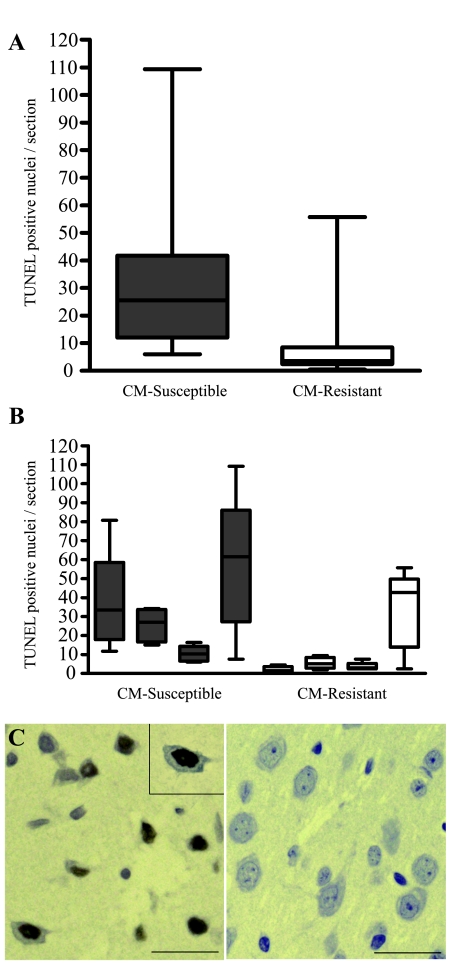
Several caspases and Fas (CD95) were prominent hubs identified in the CM interaction network (Figure 3), implicating a mechanistic role for apoptotic pathways in CM susceptibility. To confirm our computational findings, brains of uninfected and PbA-infected CM-susceptible and -resistant mice were examined for evidence of apoptosis by in situ labeling of double-stranded DNA breaks (TUNEL assay). Quantification of TUNEL-labeled cells, comparing infected CM-susceptible C57BL/6 mice to infected CM-resistant BALB/c mice, showed a significantly higher number of TUNEL-positive cells in C57BL/6 mice (Mann-Whitney test, P < 0.001) (Figure 6A). Furthermore, three of four C57BL/6 mice showed definitive TUNEL staining compared with one of four BALB/c mice (Figure 6B). Based on cellular morphology, apoptosis appeared to occur predominantly in the neurons of the cerebral cortex (Figure 6C) and in cells of the leptomeninges. One of the four C57BL/6 mice examined displayed limited cell death in the Purkinje cells (cerebellum) and brainstem in addition to the cerebral cortex.
Figure 6.
Histopathological analysis of mouse brains during PbA infection. Pathological changes in the brains of CM-susceptible (C57BL/6) and -resistant (BALB/c) mice were examined during PbA infections. Brains were formaldehyde fixed and examined using histopathology by observers blinded to the genetic background of the mice. Quantification of TUNEL-positive cells confirmed that significantly higher numbers of apoptotic cells were found in C57BL/6 mice compared with BALB/c. A: Average TUNEL-positive cells per slide were quantitated from four animals per strain (n = 20 slides per strain) and were significantly higher in C57BL/6 mice (Mann-Whitney test, P < 0.001). B: TUNEL-positive cells per slide are shown for each mouse examined. C: Representative images of TUNEL-positive neuronal cells in the cerebral cortex of an infected C57BL/6 mouse (left) and an infected BALB/c mouse (right). Scale bars represent 20 μm. TUNEL-labeled brain sections showing positive nuclear staining (brown color; inset at magnification, ×1000) suggested that apoptosis occurred in infected CM-susceptible C57BL/6 mice at day 6 after infection when compared with the equally parasitemic but CM-resistant BALB/c.
Discussion
In this study, a computational framework was developed to explore systematically the transcriptional landscape of the brain in experimental murine cerebral malaria. The salient features of expression variability were captured using principal components analysis, which showed that the dominant host response occurred in CM-susceptible mice late in the course of infection, as visualized by clustering temporal gene expression patterns (Figure 1). This response was highly enriched in processes involved in inflammation, immunity, and apoptosis (Figure 2). A gene-gene interaction network based on differentially expressed genes was constructed to help dissect the complexity of the host response and identified hubs of high connectivity, which indicate potential sites of critical transcriptional regulation (Figure 3). Many of these hubs were involved in interferon-regulated immune programs and programmed cell death. The enrichment of several interferon response elements among differentially expressed genes was independently confirmed using a computational promoter search algorithm (Figure 4).
An important mediator of apoptosis, Fas (CD95) was a dense hub within the gene-gene interactome, signifying its potential importance in cerebral malaria. Previous studies have established that Fas- and Fas-ligand (CD178)-deficient mice (lpr/lpr and gld/gld mice, respectively) are protected from CM and that Fas-mediated apoptosis may play a role in astrocyte death during the terminal stages of CM pathogenesis.44,50 The central theme of apoptosis-mediated cell death identified in the computational analysis was examined in whole-brain sections of CM-susceptible (C57BL/6) and -resistant (BALB/c) mice using TUNEL assay, which confirmed differential cerebral apoptotic responses to PbA infection (Figure 6). Previous studies examining the PbA-induced CM model suggest that the cerebral endothelium undergoes apoptosis in a perforin-dependent manner.51 Mice with symptoms of CM have also been reported to demonstrate features of neuronal52 and astrocyte53 apoptosis, which presumably occurs after blood-brain barrier disruption.53 Several in vitro studies using P. falciparum have indicated that parasitized erythrocytes induce apoptosis in mononuclear cells54,55,56 and endothelial cells.43,57 Furthermore, a small autopsy study demonstrated that 40% of CM patients examined had cleaved caspase-3 staining in their brains, indicating apoptosis, compared with 10% of patients without CM.58 Neurological deficits persist in some individuals who survive an episode of CM,59,60 and neuronal apoptosis may, in part, be responsible for these long-term cognitive effects.
Although the role of pro-inflammatory cytokines in the pathogenesis of malaria has received much attention in the literature, the precise roles of type I (eg, IFN-α and -β) and type II (IFN-γ) interferons remain unclear. Other investigators have applied transcriptional profiling to highlight the putative importance of type I and II interferon signaling in malaria infection.10,13,61 In our analysis, Ifn-γ and the interferon signaling mediators Irf-1, Stat-1, and Stat-3 all formed nodes of high connectivity in the gene-gene interaction network (Figure 3), implying that the activation of interferon-dependent transcriptional programs play a critical mechanistic role critical in the pathogenesis of CM in the PbA model. Furthermore, functional analysis indicated that major histocompatibility complex I activity is up-regulated in CM-susceptible mice, consistent with the involvement of CD8+ T-cells62 and IFN-γ production in this model. IFN-γ is required for the development of severe disease in PbA infection, because both IFN-γ-null and IFN-γ receptor-null mice do not develop CM.29,62 However, IFN-γ also aids in the control of parasitemia,29 and early production of IFN-γ, as observed in noncerebral P. berghei murine models, may be protective against progression to CM.39 Additionally, IFN-γ and IRF1 could be involved in the increased apoptosis observed in the brains of CM-susceptible (C57BL/6) mice, based on previous reports that IFN-γ induces or enhances Fas-mediated cell death in various cell types, including microglia and oligodendrocytes.63,64
A significant number of the differentially expressed transcripts identified in this study contain interferon-regulated transcription factor-binding sites, including IRF1, ISRE, and IRF7. The most significantly enriched promoter site among the differentially expressed genes was interferon-stimulated response element (ISRE), which is recognized by type I interferons via the binding of Stat1, Stat2, and Irf9.65 Genetic association studies have linked IFN-α receptor polymorphisms with protection from CM in an African population.66 In addition, IFN-α levels have been detected in PbA67 and P. falciparum infections,68 with lower levels of IFN-α observed in cases of severe malaria.41 Little is known about the relationship between type I interferon responses and malarial disease, but type I interferon responses may play a beneficial role for the host in malaria. IFN-α treatment of mice in both the P. yoelii and PbA models has been reported to improve outcome.69,70 Conversely, studies of IFN-γ treatment in P. berghei infection have yielded inconsistent results.71,72 Overall, type I and II interferon responses appear to be pivotal in the development of severe disease, and interferon therapy may represent an under-explored treatment strategy in malaria.
Host immune and metabolic responses to malaria, specifically overwhelming inflammatory responses, are important determinants of disease severity.2 This study clearly demonstrated that CM-susceptible mice develop a prominent immune-related expression profile late in infection. Presumably, this represents an overcompensated host response that not only functions to kill the parasite but also contributes to immunopathological tissue injury. In contrast, CM-resistant mice display a comparatively flat expression profile over time, with relatively minimal transcriptional response to infection altogether. Additionally, only a modest transcriptional difference was observed in CM-susceptible mice early in infection (day 1 and 3) compared with the uninfected baseline. This observation was somewhat surprising, because a previous study suggested that a surge in cytokine transcript expression occurs by day 3 after infection.39 Although CM-resistant (BALB/c) mice eventually succumb to PbA infection in the chronic stages due to hyperparasitemia and anemia,73 the lack of significant differential gene expression in the brain suggests that a diminished host response to the parasite may protect the infected host from developing CM. This observation may have important clinical implications, because delaying the onset of CM in human P. falciparum infection would allow additional time for parasite clearance with standard chemotherapy and potentially decrease the morbidity and mortality associated with this disease.
Immunomodulatory strategies that reduce overall host inflammation could delay CM onset and prove beneficial in malaria infection. By identifying key sites of transcriptional control in experimental CM, the computational approach used in the current study revealed novel targets for further investigation as possible adjuvant therapeutic strategies for malaria. As a note of caution, although PbA is a well-characterized model of CM, these findings may not all translate directly to human CM, which has pathogenic mechanisms and clinical findings distinct from murine infection. Interestingly, retinoid X receptor-α, which heterodimerizes with peroxisomal proliferator-activated receptor γ, was among the network hubs identified in the gene network analysis. The natural ligand for retinoid X receptor, 9-cis-retinoic acid, and peroxisomal proliferator-activated receptor-γ agonists, including the thiazolidinedione class of drugs, increase nonopsonic phagocytosis of P. falciparum parasitized erythrocytes and decrease tumor necrosis factor-α secretion by human monocytes.74 A second prominent hub, endothelin-1, is a potent vasoconstrictor of the cerebral microcirculation and has been implicated in the reduction of cerebral blood flow during CM in the PbA model.38 In both instances, FDA-approved drugs exist for other clinical indications that target these pathways. Specifically, a number of thiazoladinediones have been approved for treatment of diabetes mellitus, and Bosentan is an endothelin receptor antagonist approved for management of pulmonary hypertension. Although developed for treatment of clinical conditions other than malaria, these agents warrant further specific investigation as adjunctive immunomodulatory strategies to alter host response and improve clinical outcome in malaria.
Deciphering the intricate host response in cerebral malaria is a daunting task. Global assessment of this response at the genomic, transcriptomic, or proteomic levels can resolve part of its complexity and provides novel insights into its pathogenesis. An unbiased computational approach using relevant animal models of CM offers a promising framework to search for regulatory pathways of this devastating disease and identify putative targets for adjunctive therapies.
Supplementary Material
Footnotes
Address reprint requests to W. Conrad Liles, Toronto General Hospital 13E 220, 200 Elizabeth St., Toronto, ON, Canada M5G 2C4. E-mail: conrad.liles@uhn.on.ca.
See related commentary on page 1729
Supported by the Canadian Institutes of Health Research [Team Grant in Malaria CTP 79842 (K.C.K., principal investigator) and MT-13721 (to K.C.K.)], by the NIH (grant no. HL74223 to S.A.G.), by Genome Canada through the Ontario Genomics Institute, and by a Canadian Institutes of Health Research MD/PhD Studentship (to F.E.L.). K.C.K. is a Canadian Institutes of Health Research Canada Research Chair in Molecular Parasitology, and W.C.L. is a Canadian Institutes of Health Research Canada Research Chair in Infectious Diseases and Inflammation.
F.E.L. and S.A.G.contributed equally to this work. K.C.K. and W.C.L. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Severe falciparum malaria. World Health Organization, Communicable Diseases Cluster. Trans R Soc Trop Med Hyg. 2000;94(Suppl 1):S1–S90. [PubMed] [Google Scholar]
- Clark IA, Rockett KA. The cytokine theory of human cerebral malaria. Parasitol Today. 1994;10:410–412. doi: 10.1016/0169-4758(94)90237-2. [DOI] [PubMed] [Google Scholar]
- Hunt NH, Grau GE. Cytokines: accelerators and brakes in the pathogenesis of cerebral malaria. Trends Immunol. 2003;24:491–499. doi: 10.1016/s1471-4906(03)00229-1. [DOI] [PubMed] [Google Scholar]
- Adams S, Brown H, Turner G. Breaking down the blood-brain barrier: signaling a path to cerebral malaria? Trends Parasitol. 2002;18:360–366. doi: 10.1016/s1471-4922(02)02353-x. [DOI] [PubMed] [Google Scholar]
- Turner GD, Morrison H, Jones M, Davis TM, Looareesuwan S, Buley ID, Gatter KC, Newbold CI, Pukritayakamee S, Nagachinta B, White NJ, Berendt AR. An immunohistochemical study of the pathology of fatal malaria: evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- MacPherson GG, Warrell MJ, White NJ, Looareesuwan S, Warrell DA. Human cerebral malaria: a quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am J Pathol. 1985;119:385–401. [PMC free article] [PubMed] [Google Scholar]
- Brown H, Hien TT, Day N, Mai NT, Chuong LV, Chau TT, Loc PP, Phu NH, Bethell D, Farrar J, Gatter K, White N, Turner G. Evidence of blood-brain barrier dysfunction in human cerebral malaria. Neuropathol Appl Neurobiol. 1999;25:331–340. doi: 10.1046/j.1365-2990.1999.00188.x. [DOI] [PubMed] [Google Scholar]
- Lou J, Lucas R, Grau GE. Pathogenesis of cerebral malaria: recent experimental data and possible applications for humans. Clin Microbiol Rev. 2001;14:810–820. doi: 10.1128/CMR.14.4.810-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza JB, Riley EM. Cerebral malaria: the contribution of studies in animal models to our understanding of immunopathogenesis. Microbes Infect. 2002;4:291–300. doi: 10.1016/s1286-4579(02)01541-1. [DOI] [PubMed] [Google Scholar]
- Sexton AC, Good RT, Hansen DS, D’Ombrain MC, Buckingham L, Simpson K, Schofield L. Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J Infect Dis. 2004;189:1245–1256. doi: 10.1086/382596. [DOI] [PubMed] [Google Scholar]
- Hansen DS, Evans KJ, D’Ombrain MC, Bernard NJ, Sexton AC, Buckingham L, Scalzo AA, Schofield L. The natural killer complex regulates severe malarial pathogenesis and influences acquired immune responses to Plasmodium berghei ANKA. Infect Immun. 2005;73:2288–2297. doi: 10.1128/IAI.73.4.2288-2297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye NF, Coltel N, Puthier D, Flori L, Houlgatte R, Iraqi FA, Nguyen C, Grau GE, Rihet P. Gene-expression profiling discriminates between cerebral malaria (CM)-susceptible mice and CM-resistant mice. J Infect Dis. 2006;193:312–321. doi: 10.1086/498579. [DOI] [PubMed] [Google Scholar]
- Lovegrove FE, Pena-Castillo L, Mohammad N, Liles WC, Hughes TR, Kain KC. Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006;7:295. doi: 10.1186/1471-2164-7-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Batagelj V, Mrvr A. Jèunger M, Mutzel P, editors. Berlin: Springer,; Pajek-Analysis and Visualization of Large Networks. 2004 pp xii, 378. [Google Scholar]
- Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics. 2003;19:1787–1799. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun JJ, Heisler LE, Hwang II, Wilkins O, Lau SK, Hyrcza M, Jayabalasingham B, Jin J, McLaurin J, Tsao MS, Der SD. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic Acids Res. 2006;34:e85. doi: 10.1093/nar/gkl400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:REASEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman JH, Jonker RR, Keijzer R, van de Velde CJ, Cornelisse CJ, van Dierendonck JH. A new method to detect apoptosis in paraffin sections: in situ end-labeling of fragmented DNA. J Histochem Cytochem. 1993;41:7–12. doi: 10.1177/41.1.7678025. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Amani V, Vigario AM, Belnoue E, Marussig M, Fonseca L, Mazier D, Renia L. Involvement of IFN-gamma receptor-medicated signaling in pathology and anti-malarial immunity induced by Plasmodium berghei infection. Eur J Immunol. 2000;30:1646–1655. doi: 10.1002/1521-4141(200006)30:6<1646::AID-IMMU1646>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Ball HJ, MacDougall HG, McGregor IS, Hunt NH. Cyclooxygenase-2 in the pathogenesis of murine cerebral malaria. J Infect Dis. 2004;189:751–758. doi: 10.1086/381503. [DOI] [PubMed] [Google Scholar]
- Bergmann-Leitner ES, Scheiblhofer S, Weiss R, Duncan EH, Leitner WW, Chen D, Angov E, Khan F, Williams JL, Winter DB, Thalhamer J, Lyon JA, Tsokos GC. C3d binding to the circumsporozoite protein carboxy-terminus deviates immunity against malaria. Int Immunol. 2005;17:245–255. doi: 10.1093/intimm/dxh205. [DOI] [PubMed] [Google Scholar]
- Bullen DV, Hansen DS, Siomos MA, Schofield L, Alexander WS, Handman E. The lack of suppressor of cytokine signalling-1 (SOCS1) protects mice from the development of cerebral malaria caused by Plasmodium berghei ANKA. Parasite Immunol. 2003;25:113–118. doi: 10.1046/j.1365-3024.2003.00616.x. [DOI] [PubMed] [Google Scholar]
- Deininger MH, Winkler S, Kremsner PG, Meyermann R, Schluesener HJ. Angiogenic proteins in brains of patients who died with cerebral malaria. J Neuroimmunol. 2003;142:101–111. doi: 10.1016/s0165-5728(03)00250-9. [DOI] [PubMed] [Google Scholar]
- Garnica MR, de Moraes LV, Rizzo LV, de Andrade HF., Jr Supplementation of CXCL12 (CXCL12) induces homing of CD11c+ dendritic cells to the spleen and enhances control of Plasmodium berghei malaria in BALB/c mice. Immunology. 2005;115:399–406. doi: 10.1111/j.1365-2567.2005.02178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanum PS, Hayano M, Kojima S. Cytokine and chemokine responses in a cerebral malaria-susceptible or -resistant strain of mice to Plasmodium berghei ANKA infection: early chemokine expression in the brain. Int Immunol. 2003;15:633–640. doi: 10.1093/intimm/dxg065. [DOI] [PubMed] [Google Scholar]
- John CC, Opika-Opoka R, Byarugaba J, Idro R, Boivin MJ. Low levels of RANTES are associated with mortality in children with cerebral malaria. J Infect Dis. 2006;194:837–845. doi: 10.1086/506623. [DOI] [PubMed] [Google Scholar]
- Lee SH, Looareesuwan S, Chan J, Wilairatana P, Vanijanonta S, Chong SM, Chong BH. Plasma macrophage colony-stimulating factor and P-selectin levels in malaria-associated thrombocytopenia. Thromb Haemost. 1997;77:289–293. [PubMed] [Google Scholar]
- Machado FS, Desruisseaux MS, Nagajyothi , Kennan RP, Hetherington HP, Wittner M, Weiss LM, Lee SC, Scherer PE, Tsuji M, Tanowitz HB. Endothelin in a murine model of cerebral malaria. Exp Biol Med (Maywood) 2006;231:1176–1181. [PubMed] [Google Scholar]
- Mitchell AJ, Hansen AM, Hee L, Ball HJ, Potter SM, Walker JC, Hunt NH. Early cytokine production is associated with protection from murine cerebral malaria. Infect Immun. 2005;73:5645–5653. doi: 10.1128/IAI.73.9.5645-5653.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesporek S, Meyer CG, Kremsner PG, May J. Polymorphisms of transporter associated with antigen processing type 1 (TAP1), proteasome subunit beta type 9 (PSMB9) and their common promoter in African children with different manifestations of malaria. Int J Immunogenet. 2005;32:7–11. doi: 10.1111/j.1744-313X.2005.00484.x. [DOI] [PubMed] [Google Scholar]
- Luty AJ, Perkins DJ, Lell B, Schmidt-Ott R, Lehman LG, Luckner D, Greve B, Matousek P, Herbich K, Schmid D, Weinberg JB, Kremsner PG. Low interleukin-12 activity in severe Plasmodium falciparum malaria. Infect Immun. 2000;68:3909–3915. doi: 10.1128/iai.68.7.3909-3915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino P, Vouldoukis I, Dugas N, Conti M, Nitcheu J, Traore B, Danis M, Dugas B, Mazier D. Induction of the CD23/nitric oxide pathway in endothelial cells downregulates ICAM-1 expression and decreases cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell Microbiol. 2004;6:839–848. doi: 10.1111/j.1462-5822.2004.00406.x. [DOI] [PubMed] [Google Scholar]
- Pino P, Vouldoukis I, Kolb JP, Mahmoudi N, Desportes-Livage I, Bricaire F, Danis M, Dugas B, Mazier D. Plasmodium falciparum-infected erythrocyte adhesion induces caspase activation and apoptosis in human endothelial cells. J Infect Dis. 2003;187:1283–1290. doi: 10.1086/373992. [DOI] [PubMed] [Google Scholar]
- Potter SM, Chan-Ling T, Rosinova E, Ball HJ, Mitchell AJ, Hunt NH. A role for Fas-Fas ligand interactions during the late-stage neuropathological processes of experimental cerebral malaria. J Neuroimmunol. 2006;173:96–107. doi: 10.1016/j.jneuroim.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Senaldi G, Shaklee CL, Guo J, Martin L, Boone T, Mak TW, Ulich TR. Protection against the mortality associated with disease models mediated by TNF and IFN-gamma in mice lacking IFN regulatory factor-1. J Immunol. 1999;163:6820–6826. [PubMed] [Google Scholar]
- Senaldi G, Vesin C, Chang R, Grau GE, Piguet PF. Role of polymorphonuclear neutrophil leukocytes and their integrin CD11a (LFA-1) in the pathogenesis of severe murine malaria. Infect Immun. 1994;62:1144–1149. doi: 10.1128/iai.62.4.1144-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi AK, Sullivan DJ, Stins MF. Plasmodium falciparum-infected erythrocytes increase intercellular adhesion molecule 1 expression on brain endothelium through NF-kappaB. Infect Immun. 2006;74:3262–3270. doi: 10.1128/IAI.01625-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Potter S, Chaudhri G, Hansen A, Hunt NH. Fas and perforin contribute to the pathogenesis of murine cerebral malaria. Redox Rep. 1999;4:333–335. doi: 10.1179/135100099101535070. [DOI] [PubMed] [Google Scholar]
- Potter S, Chan-Ling T, Ball HJ, Mansour H, Mitchell A, Maluish L, Hunt NH. Perforin mediated apoptosis of cerebral microvascular endothelial cells during experimental cerebral malaria. Int J Parasitol. 2006;36:485–496. doi: 10.1016/j.ijpara.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wiese L, Kurtzhals JA, Penkowa M. Neuronal apoptosis, metallothionein expression and proinflammatory responses during cerebral malaria in mice. Exp Neurol. 2006;200:216–226. doi: 10.1016/j.expneurol.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Hunt NH, Golenser J, Chan-Ling T, Parekh S, Rae C, Potter S, Medana IM, Miu J, Ball HJ. Immunopathogenesis of cerebral malaria. Int J Parasitol. 2006;36:569–582. doi: 10.1016/j.ijpara.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Balde AT, Aribot G, Tall A, Spiegel A, Roussilhon C. Apoptosis modulation in mononuclear cells recovered from individuals exposed to Plasmodium falciparum infection. Parasite Immunol. 2000;22:307–318. doi: 10.1046/j.1365-3024.2000.00307.x. [DOI] [PubMed] [Google Scholar]
- Baldé AT, Sarthou JL, Roussilhon C. Acute Plasmodium falciparum infection is associated with increased percentages of apoptotic cells. Immunol Lett. 1995;46:59–62. doi: 10.1016/0165-2478(95)00017-y. [DOI] [PubMed] [Google Scholar]
- Toure-Balde A, Sarthou JL, Aribot G, Michel P, Trape JF, Rogier C, Roussilhon C. Plasmodium falciparum induces apoptosis in human mononuclear cells. Infect Immun. 1996;64:744–750. doi: 10.1128/iai.64.3.744-750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino P, Vouldoukis I, Dugas N, Hassani-Loppion G, Dugas B, Mazier D. Redox-dependent apoptosis in human endothelial cells after adhesion of Plasmodium falciparum-infected erythrocytes. Ann NY Acad Sci. 2003;1010:582–586. doi: 10.1196/annals.1299.109. [DOI] [PubMed] [Google Scholar]
- Medana IM, Mai NT, Day NP, Hien TT, Bethell D, Phu NH, Farrar J, White NJ, Turner GD. Cellular stress and injury responses in the brains of adult Vietnamese patients with fatal Plasmodium falciparum malaria. Neuropathol Appl Neurobiol. 2001;27:421–433. doi: 10.1046/j.0305-1846.2001.00360.x. [DOI] [PubMed] [Google Scholar]
- Brewster DR, Kwiatkowski D, White NJ. Neurological sequelae of cerebral malaria in children. Lancet. 1990;336:1039–1043. doi: 10.1016/0140-6736(90)92498-7. [DOI] [PubMed] [Google Scholar]
- Idro R, Ndiritu M, Ogutu B, Mithwani S, Maitland K, Berkley J, Crawley J, Fegan G, Bauni E, Peshu N, Marsh K, Neville B, Newton C. Burden, features, and outcome of neurological involvement in acute falciparum malaria in Kenyan children. JAMA. 2007;297:2232–2240. doi: 10.1001/jama.297.20.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockenhouse CF, Hu WC, Kester KE, Cummings JF, Stewart A, Heppner DG, Jedlicka AE, Scott AL, Wolfe ND, Vahey M, Burke DS. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74:5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yañez DM, Manning DD, Cooley AJ, Weidanz WP, van der Heyde HC. Participation of lymphocyte subpopulations in the pathogenesis of experimental murine cerebral malaria. J Immunol. 1996;157:1620–1624. [PubMed] [Google Scholar]
- Badie B, Schartner J, Vorpahl J, Preston K. Interferon-gamma induces apoptosis and augments the expression of Fas and Fas ligand by microglia in vitro. Exp Neurol. 2000;162:290–296. doi: 10.1006/exnr.1999.7345. [DOI] [PubMed] [Google Scholar]
- Pouly S, Becher B, Blain M, Antel JP. Interferon-gamma modulates human oligodendrocyte susceptibility to Fas-mediated apoptosis. J Neuropathol Exp Neurol. 2000;59:280–286. doi: 10.1093/jnen/59.4.280. [DOI] [PubMed] [Google Scholar]
- Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J Interferon Cytokine Res. 2002;22:835–845. doi: 10.1089/107999002760274845. [DOI] [PubMed] [Google Scholar]
- Aucan C, Walley AJ, Hennig BJ, Fitness J, Frodsham A, Zhang L, Kwiatkowski D, Hill AV. Interferon-alpha receptor-1 (IFNAR1) variants are associated with protection against cerebral malaria in the Gambia. Genes Immun. 2003;4:275–282. doi: 10.1038/sj.gene.6363962. [DOI] [PubMed] [Google Scholar]
- Huang KY, Schultz WW, Gordon FB. Interferon induced by Plasmodium berghei. Science. 1968;162:123–124. doi: 10.1126/science.162.3849.123. [DOI] [PubMed] [Google Scholar]
- Rhodes-Feuillette A, Bellosguardo M, Druilhe P, Ballet JJ, Chousterman S, Canivet M, Peries J. The interferon compartment of the immune response in human malaria: II. Presence of serum-interferon gamma following the acute attack. J Interferon Res. 1985;5:169–178. doi: 10.1089/jir.1985.5.169. [DOI] [PubMed] [Google Scholar]
- Vigário AM, Belnoue E, Cumano A, Marussig M, Miltgen F, Landau I, Mazier D, Gresser I, Renia L. Inhibition of Plasmodium yoelii blood-stage malaria by interferon alpha through the inhibition of the production of its target cell, the reticulocyte. Blood. 2001;97:3966–3971. doi: 10.1182/blood.v97.12.3966. [DOI] [PubMed] [Google Scholar]
- Vigário AM, Belnoue E, Gruner AC, Mauduit M, Kayibanda M, Deschemin JC, Marussig M, Snounou G, Mazier D, Gresser I, Renia L. Recombinant human IFN-alpha inhibits cerebral malaria and reduces parasite burden in mice. J Immunol. 2007;178:6416–6425. doi: 10.4049/jimmunol.178.10.6416. [DOI] [PubMed] [Google Scholar]
- Grau GE, Heremans H, Piguet PF, Pointaire P, Lambert PH, Billiau A, Vassalli P. Monoclonal antibody against interferon gamma can prevent experimental cerebral malaria and its associated overproduction of tumor necrosis factor. Proc Natl Acad Sci USA. 1989;86:5572–5574. doi: 10.1073/pnas.86.14.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curfs JH, van der Meide PH, Billiau A, Meuwissen JH, Eling WM. Plasmodium berghei: recombinant interferon-gamma and the development of parasitemia and cerebral lesions in malaria-infected mice. Exp Parasitol. 1993;77:212–223. doi: 10.1006/expr.1993.1078. [DOI] [PubMed] [Google Scholar]
- Neill AL, Hunt NH. Pathology of fatal and resolving Plasmodium berghei cerebral malaria in mice. Parasitology. 1992;105:165–175. doi: 10.1017/s0031182000074072. [DOI] [PubMed] [Google Scholar]
- Serghides L, Kain KC. Peroxisome proliferator-activated receptor gamma-retinoid X receptor agonists increase CD36-dependent phagocytosis of Plasmodium falciparum-parasitized erythrocytes and decrease malaria-induced TNF-alpha secretion by monocytes/macrophages. J Immunol. 2001;166:6742–6748. doi: 10.4049/jimmunol.166.11.6742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.