Abstract
Gastrointestinal disease and inflammation are common sequelae of human and simian immunodeficiency virus (SIV) infection. Nevertheless, the molecular mechanisms that lead to gastrointestinal dysfunction remain unclear. We investigated regulation of the interleukin (IL)-6-JAK-STAT3 pathway in jejunum and colon, collected at necropsy, from 10 SIV-infected macaques with diarrhea (group 1), 10 non-SIV-infected macaques with diarrhea (group 2), and 7 control uninfected macaques (group 3). All group 1 and 2 macaques had chronic diarrhea, wasting, and colitis, but group 1 animals had more frequent and severe lesions in the jejunum. A significant increase in IL-6 and SOCS-3 gene expression along with constitutive STAT3 activation was observed in the colon of all group 1 and 2 macaques and in the jejunum of only group 1 macaques compared to controls. Further, in colon, histopathology severity scores correlated significantly with IL-6 (groups 1 and 2) and SOCS-3 (group 2) gene expression. In jejunum, a similar correlation was observed only in group 1 animals. Phosphorylated STAT3 (p-STAT3) was localized to lymphocytes (CD3+) and macrophages (CD68+), with fewer CD3+ lymphocytes expressing p-STAT3 in group 1 macaques. Despite high SOCS-3 expression, STAT3 remained constitutively active, providing a possible explanation for persistent intestinal inflammation and immune activation that may favor viral replication and disease progression.
Since its initial description in 1981, human immunodeficiency virus (HIV), the causative agent of acquired immune deficiency syndrome (AIDS), has been known to cause a wide variety of illnesses by specifically targeting CD4+ T cells. Although the virus can affect essentially all organ systems, the gastrointestinal (GI) tract appears to be a major target for viral replication, CD4+ T-cell depletion, and physiological dysfunction.1,2,3 Chronic diarrhea is a very common symptom experienced by up to two-thirds of all AIDS patients at some time during the course of their disease.4,5 Although several opportunistic pathogens including protozoal, viral, bacterial, and fungal species have been implicated as contributing to diarrhea and malabsorption, the relative contributions of these agents and the possible direct contribution of HIV infection to the pathogenesis of intestinal dysfunction remains incompletely understood.6,7,8,9 These uncertainties emphasize the need to better understand the pathogenesis of intestinal dysfunction in HIV-infected individuals and develop novel therapeutic strategies to prevent the development of overt GI disease.
Simian immunodeficiency virus (SIV) infection of macaques presents a valuable model to explore the cell and molecular mechanisms that regulate virus replication in the GI tract and lead to GI inflammation and disease in HIV-infected people. The pathological changes described in the GI tract of SIV-infected macaques closely resemble those of people with HIV and AIDS.2,10,11,12,13,14,15 These include primary SIV-induced enteropathy, secondary opportunistic infections by various parasites (Cryptosporidiosis, Microsporidiosis, Trichomoniasis), viruses (cytomegalovirus, adenovirus), and bacteria (Mycobacterium avium intracellulare complex).15 Using this model, we1 and others16,17,18,19 have demonstrated an acute and profound loss of CD4+ memory T cells in the intestines of SIV-infected rhesus macaques within the first 2 weeks of infection. This finding was very important because it not only demonstrated that the intestinal tract was a preferred site of early viral replication but also provided the first significant clues linking CD4+ T-cell depletion to GI dysfunction. Similar reductions in activated CD4+ T-cell populations were recently confirmed to occur principally in the GI tract during early20 and later stages of HIV infection in humans.21 This acute loss of intestinal CD4+ T cells was recently shown to be linked to the breakdown of the intestinal barrier allowing escape of intestinal bacteria leading to significantly increased circulating levels of bacterial lipopolysaccharide in chronically HIV-infected individuals and SIV-infected rhesus macaques.22 The authors proposed this as a possible mechanism for the development of chronic immune activation facilitating viral replication and the development of AIDS.22
Although HIV and SIV do not infect structural elements (eg, epithelial cells) of the GI tract, there is abundant evidence of intestinal dysfunction, which in SIV-infected macaques begins very early in infection before any evidence of opportunistic infection.2,11,12,13,14,15 The pathogenesis of this enteropathy is not well understood, but there is ample evidence that enteropathy can be caused by intestinal immune dysfunction.23 However, the mechanisms whereby immune dysfunction caused by destruction of intestinal CD4+ T cells and immune activation (both of which occur in SIV/HIV infection) leads to disruption of intestinal function is not well understood. It is likely, however, that the close relationship between the intestinal immune system and structural components of the intestine facilitates bi-directional interactions.24 The intercellular dialogue is believed to be mediated primarily via diffusible signals such as cytokines, growth factors,25 local hormones,26,27,28 and their cognate receptors, and represents one of several complex networks of intercellular signaling pathways in the small intestine.24
Even though earlier studies have clearly indicated the role played by intestinal inflammation in the pathogenesis of GI dysfunction in HIV-infected patients29,30,31 the molecular mechanisms still remain unclear. In the GI tract, both primary HIV/SIV replication and the cytokines/chemokines elaborated thereafter by inflammatory cells have the potential to activate several signal transduction pathways. Activation of these signaling pathways secondary to virus infection may interfere with the homeostatic signaling events required to maintain normal GI structure and function, thereby leading to compromise of the intestinal barrier. Although the etiology of chronic diarrhea in HIV-infected patients remains complex, proinflammatory cytokine networks may be expected to play a central role. Up-regulation of several cytokines, such as interleukin (IL)-6, RANTES, IL-10, and interferon-γ in the gut mucosa, at least at the mRNA level, has been previously reported in HIV-infected patients.32 Among these, IL-6 is an important proinflammatory cytokine that is believed to enhance HIV replication33,34 and is also up-regulated in several chronic inflammatory conditions such as inflammatory bowel disease, rheumatoid arthritis, and numerous neoplastic conditions.35
The signal transduction pathway activated by IL-6 first involves its binding to a transmembrane receptor and subsequent activation of the Janus kinase (JAK) family of transcription factors, which then phosphorylates latent cytoplasmic proteins called STATs (signal transducer and activator of transcription), mainly STAT336,37,38 and to a lesser extent STAT1. Because excessive signaling through the JAK-STAT pathway may have deleterious consequences, the cell regulates the initiation, duration, and magnitude of the signal via a negative feedback loop involving a novel class of proteins called suppressors of cytokine signaling (SOCS).39,40 Among the seven different SOCS proteins known, SOCS-1 and SOCS-3 are known to physiologically regulate the activities of T cells and antigen-presenting cells such as macrophages and dendritic cells.41 SOCS-3, also known as SSI (STAT-induced STAT inhibitor) and cytokine-induced SH2 inhibitor (CIS3) is mainly a STAT3-induced gene and participates in the negative regulation of STAT3.42 Although it is clear that IL-6 levels are altered in the GI mucosa during HIV/SIV infection, the signal transduction pathway activated by IL-6 and its regulation in the GI tract of HIV/SIV-infected individuals remain unclear and unexplored. In the present study, using SIV-infected rhesus macaques with chronic diarrhea, we have observed constitutive activation of p-STAT3 and dysregulation of the IL-6-STAT3 signal transduction pathway in the GI tract providing a link between SIV infection and GI inflammation that could also play a role in enhancing viral replication.
Materials and Methods
Animals and Tissue Collection
Tissues were collected from a total of 27 animals including 10 animals infected with pathogenic strains of SIV (group 1) that use CCR5 in vivo and 17 animals not infected with SIV. Of the uninfected animals, 10 had chronic diarrhea (group 2) and 7 did not (group 3). The animals in group 2 with chronic nonresponsive diarrhea of no known infectious etiology have been described and used as a model of inflammatory bowel disease.43,44
It would be ideal to have a fourth group consisting of SIV-infected animals without diarrhea. Unfortunately, untreated SIV infection consistently leads to GI dysfunction and diarrhea, and hence it is not possible to include such a group. Jejunum and colon specimens were collected at necropsy from the 10 SIV-infected macaques with chronic diarrhea (group 1), 10 non-SIV-infected macaques with chronic diarrhea (group 2), and 2 uninfected control macaques (group 3) (Tables 1 and 2). In addition, pinch biopsies from jejunum and colon were collected from another five control macaques bringing the total of control macaques (group 3) used in this study to seven. Colon specimens were collected for all 27 macaques. Jejunum specimens were available for 9 of 10 group 1, 6 of 10 group 2, and 7 of 7 group 3 control macaques. All animals in groups 1 and 2 were euthanized when they became unresponsive to treatment (subcutaneous or intravenous fluids and antibiotics as appropriate based on culture and sensitivity) or lost greater than 20% of their body weight. After euthanasia with an intravenous overdose of pentobarbital, all animals received a complete necropsy and histopathological examination. All tissues were collected in RNAlater (Ambion, Austin, TX) for RNA quantification and confocal microscopy. According to the manufacturer, RNAlater protects both RNA and protein (by reversible inhibition of nucleases and proteases) in addition to preserving tissue architecture. Tissues were also collected in cryovials and snap-frozen by immersion in a 2-methylbutane/dry-ice mixture for protein extraction.
Table 1.
Animals, Inoculum, Viral Load, CD4+ T-Cell Count in Group 1 Macaques
| Animal group and no. | Duration of infection (days) | Inoculum | CD4 count cells/μl* | Plasma viral copies/ml × 106 | Viral copies/mg of total RNA × 106, Colon | Viral copies/mg of total RNA × 106, Jejunum |
|---|---|---|---|---|---|---|
| SIV-infected with diarrhea (group 1) | ||||||
| AJ82 | 232 | SIVmac251 | NA | NA | 1.95 | 0.078 |
| DD88 | 388 | SIVmac239 | 50 | 1.2 | 26.89 | 3.35 |
| CI65 | 377 | SIVmac239 | 450 | 1.5 | 4.59 | 2.5 |
| L441 | 170 | SIVmac251 and 239 | 632 | 1.26 | 15.5 | 33.2 |
| H405 | 232 | SIVmac239 | 523 | 71.4 | 17,200 | 15,480 |
| V205 | 973 | SIVmac239 | 296 | 0.3 | 8.0 | 3.45 |
| AT81 | 171 | SIVsmG932 | 640 | 0.06 | 0.5 | 0.057 |
| AT56 | 1460 | SIVmac251 and 239 | 56 | 360 | 213,000 | 16,380 |
| DT56 | 265 | SIVmac239 | 931 | 1.2 | 5.9 | NA |
| DI28 | 81 | SIVmac251 | 954 | 0.018 | 41 | 1.27 |
The lower end of the normal range for CD4+ T cells/μl of blood in the rhesus macaques is 800 cells/μl of blood.
NA, not applicable.
Table 2.
Intestinal Histopathology in Group 1, 2, and 3 Macaques
| Animal group and no. | Intestinal histopathology*
|
|
|---|---|---|
| Colon | Jejunum | |
| SIV infected with diarrhea (group 1) | ||
| AJ82 | 2 | 3 |
| DD88 | 1 and AMD | 1 and AMD |
| CI65 | 1 and AMD | 1 and AMD |
| L441 | 1 | 1 |
| H405 | 3 and CD | 2 |
| V205 | 3 | 1 and AMD, VB |
| AT81 | 3 | 3 and VB |
| AT56 | 3 and AMD, CD | 3 |
| DT56 | 3 and CD | 0 |
| DI28 | 1 | 1 |
| Non-SIV infected with diarrhea (group 2) | ||
| EI90 | 3 and CD | NA |
| EL45 | 3 and CD | NA |
| EC49 | 3 and CD | NA |
| EB12 | 3 and CD | NA |
| EM41 | 1 | 1 |
| EL71 | 3 and CD, DV | 0 |
| EB27 | 3 and CD, DV | 1 |
| DJ15 | 3 and CD | 1 |
| CT77 | 2 | 1 |
| EJ54 | 3 | 1 |
| Uninfected controls (group 3) | ||
| BV52 | 0 | 0 |
| EH70 | 0 | 0 |
| CB98 | 0 | 0 |
| CF33 | 0 | 0 |
| M302 | 0 | 0 |
| CC96 | 0 | 0 |
| R842 | 0 | 0 |
Sections of jejunum and colon were examined in a blinded manner and inflammation was scored semiquantitatively on a scale of 0 to 3 as follows: 0, within normal limits; 1, mild; 2, moderate; 3 severe. In addition, the presence of crypt dilatation (CD), villous blunting (VB), diverticulosis (DV), and amyloidosis (AMD) were recorded
NA, not applicable.
Histopathology
GI tissues were collected immediately after euthanasia and fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin (H&E) for analysis. Sections of jejunum and colon were examined in a blinded manner, and inflammation was scored semiquantitatively on a scale of 0 to 3 as follows: 0, within normal limits; 1, mild; 2, moderate; 3, severe. In addition, the presence of crypt dilatation, villous blunting, diverticulosis, and amyloidosis were recorded (Table 2).
Quantitative Real-Time SYBR Green One-Step Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Gene expression for IL-6 and SOCS-3 in the jejunum and colon was evaluated by quantitative real-time SYBR green one-step RT-PCR assay (qRT-PCR) (Qiagen Inc., Valencia, CA). Total RNA was extracted from both jejunum and colon samples using the SV total RNA isolation kit (Promega Corporation, Madison, WI), and an RNA sample representing the colon and jejunum from each macaque was assayed in triplicate wells. Each qRT-PCR reaction (25 μl) contained the following: 2× Master mix without uracil-N-glycosylase (12.5 μl), reverse transcriptase (0.25 μl), target forward and reverse primer, and total RNA (200 ng) quantified spectrophotometrically based on A260:A280 ratios. Forward and reverse primer sequence, concentration, and product size for both targets including β-actin are shown in Table 3. The PCR amplification was performed in the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, CA). Thermal cycling conditions were 50°C for 30 minutes, 95°C for 15 minutes, followed by 40 repetitive cycles of 95°C for 15 seconds, 54°C for 30 seconds, 72°C for 30 seconds. As a normalization control for RNA loading, parallel reactions in the same multiwell plate were performed using β-actin mRNA.
Table 3.
Primer Sequences Used for Real Time SYBR Green One-Step RT-PCR
| Name of gene | Primer sequence | Product size (bp) | Primer concentration |
|---|---|---|---|
| IL-6 | Forward: 5′-CCAGTACTCCCAGGAGAAGATTCCAA-3′ | 103 | 500 nmol/L |
| Reverse: 5′-CGTCGAGGATGTACCGAATGTGTT-3′ | |||
| SOCS-3 | Forward: 5′-TCTTCAGCATCTCTGTCGGAAGACC-3′ | 106 | 500 nmol/L |
| Reverse: 5′-GGCATCGTACTGGTCCAGGAACT-3′ | |||
| β-Actin | Forward: 5′-AGGCTCTCTTCCAACCTTCCTT-3′ | 108 | 300 nmol/L |
| Reverse: 5′-CGTACAGGTCTTTACGGATGTCCA-3′ |
Quantification of gene amplification after RT-PCR was made by determining the threshold cycle (CT) number for SYBR Green fluorescence within the geometric region of the semilog plot generated during PCR. Within this region of the amplification curve, each difference of one cycle is equivalent to a doubling of the amplified product of the PCR. The relative quantification of target gene expression across treatments was evaluated using the comparative CT method. The ΔCT value was determined by subtracting the β-actin CT value for each sample from the target CT value of that sample. Calculation of ΔΔCT involved using the highest sample ΔCT value (ie, sample with the lowest target expression) as an arbitrary constant to subtract from all other ΔCT sample values. Fold changes in the relative gene expression of target was determined by evaluating the expression, 2−ΔΔ CT.
Immunoprecipitation and Western Blotting
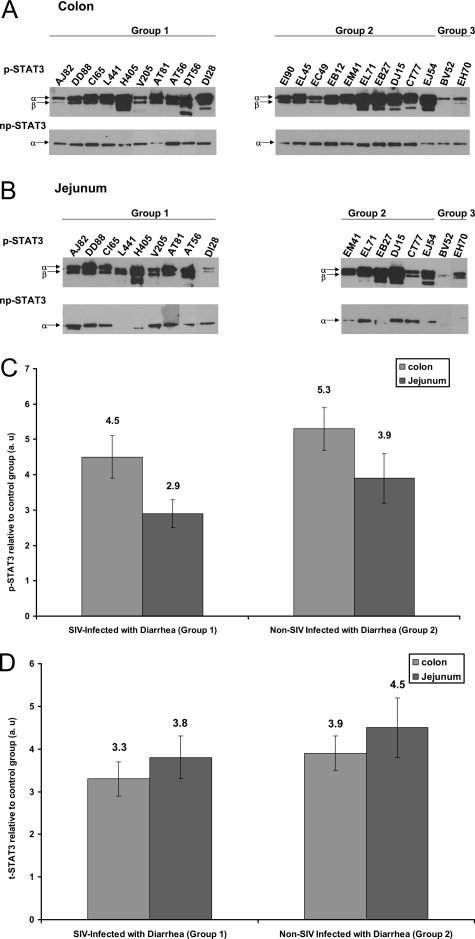
Small 2.5-cm2 pieces of colon and jejunum were crushed using a disposable polypropylene pellet pestle, and protein extraction was performed in a lysis buffer (Cell Signaling Technology, Inc., Beverly, MA) containing 20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L Na2EDTA, 1 mmol/L EGTA, 1% Triton, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 μg/ml leupeptin, protease inhibitor cocktail, and phosphatase inhibitor cocktail (Sigma Chemical Company, St. Louis, MO). Of the seven group 3 macaques, protein lysates prepared from only two macaques (BV52 and EH70) were used for Western blotting and immunoprecipitation. Approximately, 500 μg of total protein extract was first precleared with 4 μl of normal mouse immunoglobulin for ∼2 hours and then immunoprecipitated with ∼3.5 μl of a mouse monoclonal antibody against p-STAT3 (Cell Signaling Technology), overnight at 4°C followed by incubation with 25 μl (50% w/v) of protein G agarose beads (Invitrogen Corp., Carlsbad, CA) at 4°C for 4 to 5 hours. The supernatant was removed and transferred to a separate 1.5-ml microcentrifuge tube, and any remaining nonphosphorylated STAT3 (np-STAT3) was immunoprecipitated using a rabbit polyclonal antibody (∼4 μl) (Santa Cruz Biotechnology, Santa Cruz, CA) raised against total STAT3 (t-STAT3) at 4°C overnight on a shaker. Immunoprecipitated p-STAT3 and np-STAT3 proteins were heat denatured for 5 minutes at 100°C in sample loading buffer containing 62.5 mmol/L Tris-HCl, 5% 2-mercaptoethanol, 10% glycerol, 2% sodium dodecyl sulfate (SDS), and bromophenol blue, resolved on 8% SDS-polyacrylamide gel electrophoresis (PAGE) gels and transferred to 0.45-μm nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA). The membranes were probed with a rabbit polyclonal primary antibody against p-STAT3 (Cell Signaling Technology) and mouse monoclonal antibody against t-STAT3 (Cell Signaling Technology) followed by the appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Membranes were treated with Supersignal West-Pico chemiluminescent substrate (Pierce Biotechnology Inc., Rockford, IL) for 5 minutes, and the signal was developed by exposing the membrane to X-OMAT Kodak film (Eastman Kodak, Rochester, NY) for 10 minutes for both p-STAT3 and t-STAT3. Since the tissues were collected throughout a period of 2 years, immunoprecipitation and Western blotting were performed separately as and when the tissues were available. Hence the images shown in Figure 2 were cropped, aligned, and fused into one composite image.
Figure 2.
IL-6-mediated STAT-3 activation at tyrosine 705 occurs in the colon (A) and jejunum (B) of SIV-infected (group 1) and non-SIV-infected macaques (group 2) with diarrhea. p-STAT3 and then np-STAT3 were sequentially immunoprecipitated from the same lysates of colon and jejunum and run on SDS-PAGE, and thereafter, the membrane was probed with appropriate antibodies. Two control macaques (group 3) (BV52 and EH70) were used for the STAT3 Western analysis. Because the tissues were collected throughout a period of 2 years, immunoprecipitation and Western blotting were performed separately as the tissues were obtained. Hence the images shown in A and B were cropped, aligned, and merged into one composite figure. Quantitative densitometric analysis of p-STAT3 and t-STAT3 (p-STAT3 + np-STAT3 = t-STAT3) protein expression are shown in C and D, respectively, for the colon and jejunum of groups 1 and 2 macaques relative to group 3 (control animals). Values are expressed in arbitrary units (a.u.). The relative fold increases for individual animals in both groups was summed and averaged to obtain a single figure that is shown on top of each bar graph. The error bars represent average fold difference for each group ± SEM.
Biotin Streptavidin Pull Down Assay
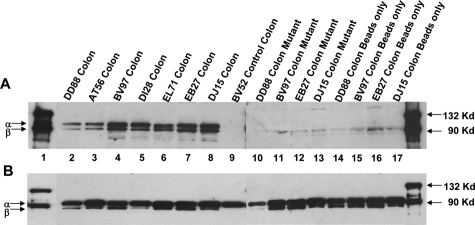
Two oligonucleotides, one corresponding to the STAT3 consensus binding site sense-biotin-5′-GATCCTTCTGGGCCGTCCTAGATC-3′ and another mutant oligonucleotide sense-biotin-5′-GATCCTTCTGGGAATTCCTAGATC-3′ with an AAT to CCG substitution in the STAT3 binding motif (underlined) were used in the pull down assays according to the protocol described by Ragione and colleagues45 with minor modifications. Both oligonucleotides contained a biotin molecule conjugated to the nucleotide at the 5′-position. The sense and antisense (not shown) strands of both wild-type and mutant oligonucleotides were annealed in vitro to generate 24-bp double-stranded DNA fragments. The annealed oligonucleotides were first separated on a 5% metaphor high-resolution agarose gel (Cambrex Corporation, East Rutherford, NJ), after which the bands were cut and the 24-bp DNA purified using the QIAEX-2 gel extraction kit (Qiagen Inc.). Total protein was extracted as previously described. Approximately 500 μg of total protein extracted from colon samples was incubated with 1 μg of either wild-type or mutant STAT3 oligonucleotide for ∼20 minutes at room temperature in a binding buffer containing 10 mmol/L Tris (pH 7.5), 150 mmol/L KCl, 1 mmol/L EDTA, 10 mmol/L CaCl2, 5 mmol/L MgCl2, and 1 mmol/L dithiothreitol. Subsequent to the incubation, 25 μl of streptavidin agarose beads (50% w/v) (Invitrogen Corp.) was added, and the tubes were incubated for 2 hours at 4°C. After the incubation, the tubes were centrifuged, and the supernatant was transferred to a separate 1.5-ml microcentrifuge tube. The lysate was then immunoprecipitated overnight for t-STAT3 (includes p-STAT3 and np-STAT3) as described earlier. The beads were washed once with binding buffer, resuspended in 20 μl of sample loading buffer containing 62.5 mmol/L Tris-HCl, 5% 2-mercaptoethanol, 10% glycerol, 2% SDS, and bromophenol blue and heat denatured at 100°C for ∼5 minutes. The tubes were briefly centrifuged, and the supernatant was loaded on a 8% SDS-polyacrylamide gel. Detection of p-STAT3 was performed as described under immunoprecipitation and Western blotting with minor modifications to the procedure described earlier. The membrane shown in Figure 4A after incubation with a rabbit polyclonal antibody against p-STAT3 (1 in 1000) (Cell Signaling) was treated with Supersignal West-Femto chemiluminescent substrate (Pierce Biotechnologies) for 5 minutes, and the signal was developed by exposing the membrane to X-OMAT Kodak film for 10 seconds. Similarly, the membrane shown in Figure 4B was incubated with a mouse monoclonal antibody against t-STAT3 (1 in 2000) (Cell Signaling) and was later treated with Supersignal West-Pico chemiluminescent substrate (Pierce Biotechnologies) for 5 minutes, and the signal was developed by exposing the membrane to X-OMAT Kodak film for 5 minutes.
Figure 4.
Protein extracts of colon prepared from four SIV-infected macaques with chronic diarrhea (lanes 2 to 5, 10 and 11, 14 and 15), three non-SIV-infected macaques with diarrhea (lanes 6 to 8, 12 and 13, 16 and 17) and one control macaque (lane 9) were incubated with biotin-labeled wild-type STAT3-binding oligonucleotide sequence (lanes 2 to 9) or mutant (lanes 10 to 13) or streptavidin-conjugated agarose beads only (lanes 14 to 17). The protein-DNA complexes were recovered as described in the Materials and Methods and analyzed by immunoblotting using antibodies against p-STAT3 (A) and t-STAT3 (B). Thus, A shows the presence or absence of p-STAT3 and B shows the presence of t-STAT3 (p-STAT3 and np-STAT3) in the same sample shown in A.
Confocal Microscopy
To determine the cell types in the GI tract expressing p-STAT3, we performed double-label confocal microscopy. Tissues collected in RNAlater were first fixed in 2% paraformaldehyde for 4 hours, cryopreserved overnight in 30% sucrose solution, and embedded in Tissue Tek OCT (optimal cutting temperature compound; Miles Inc., Elkhart, IN). Embedded tissues were sectioned at 10 μm and stained with the appropriate primary and secondary antibodies. Briefly, slides were blocked with 100 μl of blocking buffer (10 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 3% bovine serum albumin, 10% normal goat serum, and 0.1% Triton X-100) for 1 hour followed by overnight incubation at 4°C with rabbit polyclonal p-STAT3 antibody (1:200 dilution) (Cell Signaling Technology). The slides were washed three times in buffer (10 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, and 0.1% Triton X-100) followed by addition of goat anti-rabbit secondary antibody conjugated to Alexa 568 (1:1000) (Invitrogen Corp.). This was followed by mouse anti-CD68 (IgG1 macrophage clone KP1 at 1:20) or mouse anti-CD3 (IgG1 clone F 7.2.38 at 1:20) at room temperature for 1 hour. Both antibodies were obtained from DAKO, Carpinteria, CA. The slides were washed three times and incubated for 1 hour with goat anti-mouse secondary antibody conjugated with Alexa 488 (1:1000). Confocal microscopy was performed using a Leica TCS SP2 confocal microscope equipped with three lasers (Leica Microsystems, Exton, PA). Individual optical slices represent 0.2 μm, and 32 to 62 optical slices were collected at 512 × 512 pixel resolution. NIH Image (version 1.62; National Institutes of Health, Bethesda, MD) and Adobe Photoshop (version 7.0; Adobe Systems, San Jose, CA) were used to assign colors to the three channels collected: Alexa 568 is red, Alexa 488 (Invitrogen Corp) is green, and the differential interference contrast image is in gray scale. The three channels were collected simultaneously. Co-localization of antigens is demonstrated by the addition of colors as indicated in the figure legends.
Quantitation of Mucosal Viral Loads
Total RNA samples from all group 1 animals were subjected to a quantitative real-time TaqMan two-step RT-PCR analyses to determine the viral load in SIV-infected colon and jejunum samples. Briefly, primers and probes specific to the SIV LTR sequence were designed and used in the real-time TaqMan PCR assay. Probes were conjugated with a fluorescent reporter dye (FAM) at the 5′ end and a quencher dye at the 3′ end. Fluorescence signal was detected with an ABI Prism 7700 sequence detector (PE Applied Biosystems). Data were captured and analyzed with Sequence Detector Software (PE Applied Biosystems). Viral copy number was determined by plotting CT values obtained from the colon and jejunum samples against a standard curve (y = −3.351x + 40.377) (r2 = 0.998) generated with in vitro-transcribed RNA representing known viral copy numbers. Plasma viral loads for 9 of 10 group 1 macaques were performed using the b-DNA assay for SIV viral RNA (Bayer Diagnostics, Tarrytown, NY).
Statistical Analysis
Gene expression for IL-6 and SOCS-3 among groups was compared using the Kruskal-Wallis one-way analysis of variance by ranks for nonparametric independent group comparisons. When the results of Kruskal-Wallis one-way analysis of variance by ranks indicated a significant difference at the P < 0.001 level between the groups examined, Dunn’s multiple comparison procedure was then used to identify the groups that differed at the P < 0.05 significance level. Histopathology severity scores for jejunum and colon in group 1, 2, and 3 animals were analyzed using Kruskal-Wallis one-way analysis of variance by ranks for nonparametric independent group comparisons. Within each group histopathology severity scores between colon and jejunum were analyzed using Dunn’s multiple comparison procedure to identify the groups that differed at the P < 0.05 significance level. The presence of other histopathological lesions such as villous blunting, crypt dilatation, amyloidosis in the jejunum and colon of group 1 and 2 animals were analyzed using two-tailed Fisher exact test. A correlation coefficient was also calculated to assess the degree of association between histopathology severity scores and gene expression for IL-6 and SOCS-3 in the colon and jejunum of both groups.
Results
Intestinal Histopathology
Histological evaluation of H&E-stained sections of colon and jejunum (Table 2) revealed the presence of enterocolitis in most animals in groups 1 and 2 but not in group 3. When examining colon and jejunum separately, there did not appear to be much difference between groups 1 and 2 with regard to the presence of colitis (10 of 10 group 1 animals and 10 of 10 group 2 animals). However, the severity of the inflammation based on semiquantitative histopathological grading criteria and blinded analysis by a board certified pathologist (A.A.L.) suggested that differences do exist (Table 4 and Supplementary Figure 1, see http://ajp.amjpathol.org). Specifically, the inflammation in the GI tract of group 1 animals was fairly uniform in the colon and jejunum whereas in group 2 animals the colon was much more severely affected (P = 0.004) (Tables 2 and 3). These differences, based primarily on the extent and severity of the inflammatory infiltrates, were mirrored by the incidence of other intestinal lesions (Table 4) such as villous blunting, crypt dilatation, and abscess and amyloidosis in the jejunum of group 1 and not in group 2 animals. However, the presence of other intestinal lesions in the jejunum of group 1 macaques lacked statistical significance. Opportunistic pathogens such as cytomegalovirus and Mycobacterium avium intracellulare complex were detected in only 3 of 10 group 1 macaques (Table 5). Campylobacter coli, Campylobackter jejuni, or Shigella flexneri were isolated from the colon at necropsy in 2 of 10 group 1 and 5 of 10 group 2 macaques (Table 5).
Table 4.
Comparison and Statistical Significance of Histopathological Lesions
| Tissue | Animal and score details | Group 1 | Group 2 | Group 3 | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| G1 versus G2 | G1 versus G3 | G2 versus G3 | Colon versus Jej − G1 | Colon versus Jej − G2 | |||||
| Colon | No of animals with colitis/total number of animals examined | 10/10 | 10/10 | 0/7 | NSD | <0.005 | <0.0005 | NSD | = 0.004 |
| Total scores | 21 | 27 | 0 | ||||||
| Average scores | 2.1 | 2.7 | 0 | ||||||
| Number of animals with other lesions/total number of animals examined* | 5/10 | 4/10 | NA | NSD | NA | NA | NSD | NSD | |
| Jejunum | No of animals with enteritis/total number of animals examined | 9/9 | 5/6 | 0/7 | NSD | <0.0008 | <0.005 | ||
| Total scores | 16 | 5 | 0 | ||||||
| Average scores | 1.78 | 0.83 | 0 | ||||||
| Number of animals with other lesions/total number of animals examined* | 4/10 | 0/6 | NA | NSD | NA | NA | |||
NSD, not significantly different; NA, not applicable.
Lesions such as villous blunting, crypt dilation/abscesses, amyloidosis, diverticulosis, and so forth.
Table 5.
Opportunistic Pathogens and Intestinal Bacterial Isolates
| Duration of infection (days) | OI, small/large intestine | Other OI | Bacterial isolates* | |
|---|---|---|---|---|
| SIV-infected with diarrhea (group 1) | ||||
| AJ82 | 232 | CMV inclusion bodies, colon | None | C. coli |
| DD88 | 388 | M. avium intracellulare | None | None |
| CI65 | 377 | M. avium intracellulare | None | C. coli |
| L441 | 170 | None | P. carinnii, lung | None |
| H405 | 232 | None | None | None |
| V205 | 973 | None | None | None |
| AT81 | 171 | None | None | None |
| AT56 | 1460 | None | C. albincans, tongue/esophagus | None |
| DT56 | 265 | None | P. carinnii, lung | None |
| DI28 | 81 | None | None | None |
| Non-SIV infected with diarrhea (group 2) | ||||
| EI90 | NA | None | None | C. jejuni |
| EL45 | NA | None | None | C. coli |
| EC49 | NA | None | None | None |
| EB12 | NA | None | None | None |
| EM41 | NA | None | None | C. coli |
| EL71 | NA | None | None | None |
| EB27 | NA | None | None | S. flexneri, C. coli |
| DJ15 | NA | None | None | None |
| CT77 | NA | None | None | C. coli |
| EJ54 | NA | None | None | None |
| Uninfected control (group 3) | ||||
| BV52 | NA | None | None | None |
| EH70 | NA | None | None | None |
| CB98 | NA | None | None | None |
| CF33 | NA | None | None | None |
| M302 | NA | None | None | None |
| CC96 | Alive | None | None | None |
| R842 | NA | None | None | None |
OI, opportunistic infection; NA, not applicable.
All from colon at necropsy, no small intestinal isolates obtained.
Plasma and Mucosal Viral Loads and CD4+ T Cell Counts
The viral loads in the plasma, colon, and jejunum of all of the SIV-infected macaques (group 1) are shown in Table 1 and were obtained at necropsy. All SIV-infected macaques (group 1) had substantial mucosal and plasma viral loads. Viral loads in the jejunum had a range from 0.057 × 106 to 16,380 × 106 copies/mg total RNA with a median of 3.35 × 106 copies/mg total RNA. The colon generally had higher viral loads than the jejunum for the same animal with a range from 0.5 × 106 to 213,000 × 106 copies/mg total RNA with a median of 11.75 × 106] copies/mg total RNA. Peripheral CD4+ T-cell counts obtained at necropsy were available for 9 of 10 group 1 macaques (Table 1). Of the 10 group 1 macaques, at least 7 had low CD4+ T-cell counts.
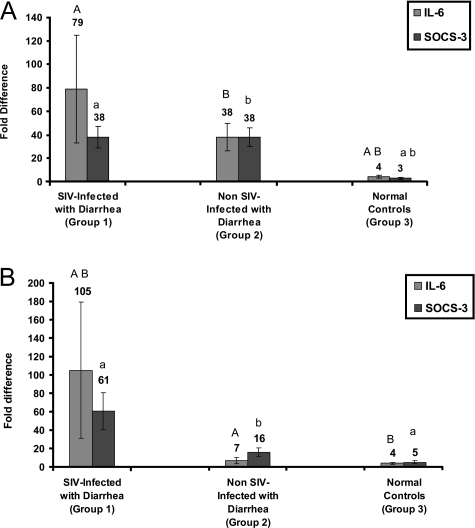
Mucosal Gene Expression for IL-6 and SOCS-3
To verify that the occurrence of diarrhea in SIV-infected macaques was associated with a marked expression of IL-6, qRT-PCR was performed using total RNA extracted from jejunal and colonic mucosal samples from all macaques. Individual fold changes in gene expression calculated as described in Materials and Methods for IL-6 and SOCS-3 in all three groups are shown in Table 6. Figure 1, A and B, shows averaged group wise fold differences for IL-6 and SOCS-3, in the colon and jejunum, respectively. In the colon, IL-6 gene expression in group 1 and group 2 animals was significantly increased compared to normal controls (P < 0.05) (Figure 1A). Further, a statistically significant correlation was also observed between histopathology severity scores and IL-6 gene expression in group 1 [r2 = 0.6 (P < 0.0005)] and group 2 [r2 = 0.67 (0.025 < P < 0.01)] macaques (Tables 2, 4, and 6). In the jejunum, IL-6 gene expression in group 1 macaques differed significantly from both group 2 and normal controls (Figure 1B).
Table 6.
Individual Fold Differences in IL-6 and SOCS-3 Gene Expression
| Group | Animal no. | IL-6*
|
SOCS-3*
|
||
|---|---|---|---|---|---|
| Colon | Jejunum | Colon | Jejunum | ||
| SIV-infected with diarrhea (group 1) | AJ82 | 11 | 27 | 17 | 39 |
| DD88 | 46 | 696 | 31 | 135 | |
| CI65 | 16 | 72 | 28 | 42 | |
| L441 | 5 | 3 | 22 | 4 | |
| H405 | 77 | 12 | 12 | 13 | |
| V205 | 14 | 17 | 12 | 32 | |
| AT81 | 482 | 71 | 80 | 121 | |
| AT56 | 107 | 43 | 98 | 153 | |
| DT56 | 24 | NA | 52 | NA | |
| DI28 | 7 | 4 | 21 | 7 | |
| Non-SIV infected with diarrhea (group 2) | EI90 | 27 | NA | 38 | NA |
| EL45 | 26 | NA | 20 | NA | |
| EC49 | 28 | NA | 34 | NA | |
| EB12 | 55 | NA | 57 | NA | |
| EM41 | 8 | 2 | 8 | 3 | |
| EL71 | 134 | 1 | 97 | 7 | |
| EB27 | 21 | 12 | 20 | 25 | |
| DJ15 | 16 | 2 | 52 | 4 | |
| CT77 | 12 | 21 | 30 | 25 | |
| EJ54 | 51 | 3 | 24 | 29 | |
| Control animals (group 3) | BV52 | 2 | 6 | 2 | 13 |
| CB98 | 5 | 3 | 7 | 4 | |
| CF33 | 9 | 8 | 3 | 7 | |
| M302 | 2 | 2 | 2 | 1 | |
| CC96 | 2 | 2 | 1 | 2 | |
| R842 | 1 | 2 | 4 | 3 | |
Fold changes in the relative gene expression were determined by evaluating the expression, 2−DΔ CT.
Colon and jejunum specimens from EH70 (group 3) were not used for qRT-PCR analysis.
NA, not applicable.
Figure 1.
Relative abundance in gene expression for IL-6 and SOCS-3 detected using quantitative real-time SYBR Green one-step RT-PCR. A and B represent colon and jejunum, respectively, from SIV-infected animals with diarrhea, non-SIV-infected with diarrhea, and uninfected control macaques. The fold differences in gene expression were calculated as described in Materials and Methods. The relative fold increases for each group was summed and averaged to obtain a single figure that is shown on top of each bar graph. The error bars represent average fold difference for each group ± SEM. Bar graphs with same letter and case (eg, A and A) differ significantly (P < 0.05). The upper case letters denoted above gray bars represent IL-6. Similarly, the lower case letters shown above black bars represent SOCS-3.
In response to IL-6, activation of STAT3 should induce the expression of SOCS-3, which in turn should negatively regulate IL-6 signaling. As shown in Figure 1A, SOCS-3 gene expression in the colon of animals with diarrhea differed significantly from normal control macaques (P < 0.05). The magnitude of SOCS-3 expression appears similar in the colon of animals with diarrhea in comparison to controls regardless of whether they are infected with SIV or not. Interestingly, a strong correlation between histopathology severity scores (Table 4) and SOCS-3 gene expression was observed only in the colon of group 2 macaques (r2 = 0.67) (0.025 < P < 0.001) (Tables 2, 4, and 6). In contrast, in the jejunum, gene expression for SOCS-3 in group 1 macaques was found to be significantly different from normal controls (P < 0.05) (Figure 1B) and showed a strong correlation with histopathology severity scores (r2 = 0.61) (0.1 < P < 0.05) (Tables 2 and 6). On the other hand, SOCS-3 gene expression in the jejunum of group 2 was not significantly different from group 1 or from normal controls. This is probably related to the fact that the majority of group 1 macaques had moderate to severe inflammation of the jejunum and colon (enterocolitis), whereas in group 2 animals the colon was more severely affected with minimal involvement of the jejunum based on histopathology (Table 2). The statistically significant increase in IL-6 in the jejunum of group 1 versus group 2 animals further supports this.
STAT3 Is Activated in the Intestine of Macaques with Diarrhea Regardless of Infection with SIV
To determine whether IL-6 was in fact exerting proinflammatory effects on cells at the site of inflammation, we investigated the activation status of STAT3 molecules, which occurs by phosphorylation at Tyr 705. Substantial amounts of p-STAT3 isoforms (p-STAT3α, 86 kDa; and p-STAT3β, 79 kDa) were detected in the colon of all group 1 and 2 macaques (Figure 2A). In contrast, a weak p-STAT3 band was detected in the colon and jejunum of group 3 macaques (BV52 and EH70) (Figure 2, A and B). In the jejunum, with the exception of two animals (L441 and DI28), significant amounts of p-STAT3 were observed compared to the control macaques (Figure 2B). In two animals (DT56 colon and H405 jejunum), a third band was seen and is mainly attributable to minimal protein degradation. Even though IL-6 and SOCS-3 gene expression in the jejunum of group 2 macaques were not statistically different from controls (Figure 1B), we did see significant amounts of p-STAT3 protein in at least five of six animals (Figure 2B). Further, densitometric analysis performed on the blots showed clear up-regulation of p-STAT3 in the jejunum of group 1 (∼2.9-fold) and 2 (∼3.9-fold) macaques compared to group 3 (Figure 2C). The band intensities for np-STAT3 protein in the colon and jejunum of group 1 and group 2 macaques were generally weak compared to p-STAT3. Because the same protein lysate sample was first used for immunoprecipitating p-STAT3 followed by np-STAT3, these findings would suggest that the STAT3 proteins in the colon and jejunum of both group 1 and group 2 macaques used in this study are predominantly phosphorylated (p-STAT3 band > np-STAT3 band) and hence in a constitutively activated state. Further, essentially all of the STAT3β (79 kDa) isoform appears to be phosphorylated because this isoform was not detected in the np-STAT3 fraction using the t-STAT3 antibody (Figure 2A). Finally, the densitometric analysis also suggests that, on the whole, there is considerably increased amounts of t-STAT3 in the colon and jejunum of group 1 and 2 macaques, and a greater percentage of it is phosphorylated compared to controls (Figure 2, C and D).
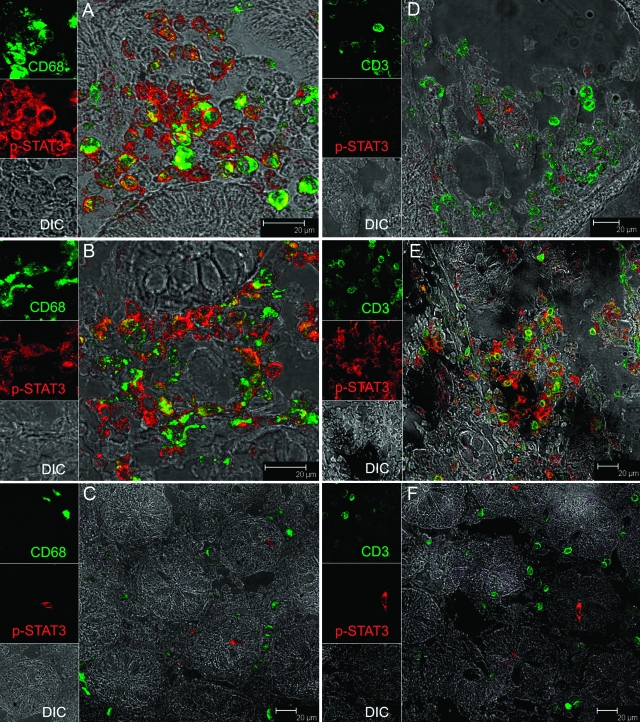
Lamina Propria Mononuclear Cells Express High Levels of STAT-3 in the Colon of SIV-Infected and Non-SIV-Infected Macaques with Diarrhea
To identify the cell types that express p-STAT3, we performed confocal microscopy analysis of colon tissue from three group 1 macaques (AT56, H405, D128), two group 2 (DJ15, EL45) macaques, and one group 3 (BV52) macaque with representative images for each group presented in Figure 3 and Supplementary Figure 2 (see http://ajp.amjpathol.org). In all three SIV-infected macaques, macrophages expressing CD68 were the primary cell type that expressed p-STAT3 (Figure 3A). A minority of T cells in the lamina propria expressing CD3 were also found to express p-STAT3 (Figure 3D). In contrast, in the non-SIV-infected macaques with diarrhea, we observed large numbers of both CD68+ (Figure 3B) and CD3+ cells expressing p-STAT3 (Figure 3E). p-STAT3 was mainly localized to the cytoplasm of macrophages and lymphocytes with very few cells exhibiting nuclear localization. In contrast to animals with chronic diarrhea, in normal controls (BV52) few cells were positive for p-STAT3, and they were neither CD68+ nor CD3+ (Figure 3, C and F).
Figure 3.
Immunophenotype of p-STAT3-positive cells in the colon of SIV-infected macaques with diarrhea (A and D), non-SIV-infected macaque with diarrhea (B and E), and an uninfected control macaque (C and F). All panels involve double labels with p-STAT3 (red) and either CD68 for macrophages (A–C) or CD3 for T cells (D–F), which appear green. For each panel the individual channels [green for CD3 or CD68, red for p-STAT3, and gray for differential interference contrast to reveal tissue architecture] appear on the left with a larger merged image on the right. Co-localization of labels appears yellow. Note that in SIV-infected animals with diarrhea (A and D) most cells positive for p-STAT3 are CD68+ macrophages whereas in non-SIV-infected animals with diarrhea (B and E) both CD68+ macrophages and CD3+ T cells are p-STAT3+. Interestingly, in the normal control animal (C and F), not only are p-STAT3+ cells rare in comparison, but they are also negative for both CD3 and CD68. Images shown are representative of three group 1 macaques, two group 2 macaques, and a control animal and were obtained from H405 (A and D), DJ15 (B and E) and BV52 (C and F).
DNA Binding Activity of STAT3 Is Increased in the Colon of SIV-Infected Macaques and Non-SIV-Infected Macaques with Diarrhea
After phosphorylation at Tyr 705, STAT3 translocates to the nucleus where it binds to specific sequence elements present in the promoter regions of various STAT-responsive genes. However, confocal microscopy demonstrated STAT3 to be cytoplasmic rather than nuclear in majority of the cells. Therefore, we next investigated the DNA binding activity of STAT3 using the biotin streptavidin pull-down assay to determine whether these activated STAT3 proteins were biologically active. In this experiment, using the double-stranded STAT3-binding sequence as bait, we observed that STAT3 was not only phosphorylated but also had the capacity to bind DNA. As shown in Figure 4, the STAT3 oligonucleotide specifically precipitated a considerable amount of p-STAT3 protein from the colon of four group 1 and three group 2 macaques used in this experiment (Figure 4A). No p-STAT3 protein was precipitated by the STAT3 oligonucleotide from the control group 3 macaque (BV52) (Figure 4A), mainly attributable to the lack of availability of substantial amounts of the biologically active form of STAT3 (p-STAT3), which is also evident in Figure 2A. The specificity of the binding is shown by the fact that the mutated oligonucleotide failed to precipitate any activated STAT3 protein (Figure 4A). t-STAT3 (p-STAT3 and np-STAT3) was present in all lanes (Figure 4B) and points to the fact that the absence of the p-STAT3 signal (Figure 4A) in lanes 9 to 17 was not attributable to the complete absence of STAT3 in the lysate. Thus the absence of a p-STAT3 band in lanes 9 to 17 in Figure 4A is attributable to the lack of up-regulation of p-STAT3 in the control (lane 9) or mutation in the STAT3-binding site in lanes 10 to 13 or absence of STAT3 oligonucleotide in lanes 14 to 17.
Discussion
The GI tract is a major reservoir of CCR5+ CD4+ T lymphocytes of the memory phenotype making it a major target for infection with HIV/SIV.1,19,46 Recent findings in humans further strengthens the notion that HIV/SIV replication occurs predominantly in the gut and other mucosal lymphoid tissues.20,21,47 Up to two-thirds of HIV-infected patients and essentially all untreated SIV-infected macaques develop symptoms of GI dysfunction such as diarrhea during the course of infection. Although several causative factors have been proposed, the ensuing molecular pathological mechanisms that initiate GI disease after massive CD4+ T-cell loss still remain unclear. Subsequent to HIV/SIV infection, up-regulation of several proinflammatory cytokines (eg, IL-1β, IL-6, IL-8, interferons, and tumor necrosis factor-α) occurs in the GI tract, which may promote GI inflammation.31,32 IL-6 is an important proinflammatory cytokine that uses the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway for signal transduction.36,37,38 Although several studies in the past have attempted to characterize HIV/SIV-associated mucosal cytokine expression, there is very little information regarding the downstream molecules that these cytokines activate, especially in the GI tract of patients experiencing chronic diarrhea and weight loss. Therefore, in the present study, jejunal and colon specimens from SIV and non-SIV-infected rhesus macaques with weight loss and diarrhea were examined to assess if the activation status of the JAK/STAT pathway could be linked to the development of GI inflammation and disease. In the present study, we demonstrate that STAT3, activated as a result of proinflammatory cytokine signaling in the GI tract of SIV-infected and noninfected rhesus macaques may be an important mediator of inflammation resulting in chronic diarrhea and wasting. Although alterations in the signaling pathways in the colon were similar regardless of whether the animals were SIV infected, the jejunum was more significantly impacted in SIV-infected animals with diarrhea with significant up-regulation of IL-6 and SOCS-3 as well as more severe histopathological lesions compared to the other groups. This is consistent with the dramatic impact SIV has on the entire GI tract. Furthermore, the presence of high STAT3 activation despite high levels of SOCS-3 suggests dysregulation of the JAK-STAT pathway in animals with chronic diarrhea. In the context of SIV infection, this would favor continuing inflammation and immune activation, which would favor viral replication and disease progression.
IL-6 has been described to be a multifunctional cytokine because it is known to regulate several processes including the inflammatory response.48 Almost a decade ago, Snijders and colleagues49 observed no evidence of cytokine-mediated inflammation in homogenized jejunal mucosa of HIV-infected individuals. However, in the same study, after culture, IL-6 concentrations were higher in HIV-infected patients with diarrhea than in HIV-negative controls, although the results were not statistically significant. Most recent studies suggest a strong association between HIV infection and mucosal inflammation31,50 and that cytokines, such as IL-6, RANTES, IL-10, and interferon-γ, belonging to both Th1 and Th2 types are significantly up-regulated.32 Specifically with regard to IL-6, McGowan and colleagues32 observed increased IL-6 mRNA expression in the GI mucosa of all HIV-infected patients regardless of the mucosal viral load. In the present study, we observed several fold increases in IL-6 mRNA expression in both colon and jejunum of SIV-infected macaques with diarrhea (group 1) compared to controls (group 3). Interestingly, although enhanced IL-6 gene expression was also observed in the colon of non-SIV-infected macaques with diarrhea (group 2), there was no significant increase in IL-6 in the jejunum of these animals compared to controls. This finding is again substantiated by the significant correlation detected between histopathology severity scores and gene expression for IL-6 only in the colon and not jejunum of group 2 animals. Furthermore, IL-6 mRNA expression was significantly elevated in the jejunum of SIV-infected macaques with diarrhea (group 1) compared to non-SIV-infected macaques with diarrhea (group 2). In support of this, we found increased viral loads in the colon and jejunum of group 1 macaques occurring together with high IL-6 gene expression. Collectively, these data suggest that the generalized immune activation associated with HIV/SIV infection is driving the increase in IL-6 throughout the GI tract of SIV-infected animals (group 1).
The presence of IL-6 mRNA in the present and previous studies does not prove that it exerts receptor-mediated intracellular signaling. In contrast to alterations in cytokine synthesis, posttranslational modifications such as phosphorylation and dephosphorylation involving STAT proteins in response to cytokine signaling is a rapid event lasting only a few minutes. In the present study, using two independent procedures, we found that the transcription factor STAT3 undergoes posttranslational changes such as phosphorylation, possesses enhanced DNA binding activity, and remains constitutively active in the colon of SIV-infected (group 1) and non-SIV-infected macaques (group 2) with diarrhea compared to the normal control macaques (group 3). Significant amounts of activated STAT3 were also observed in the jejunum of, at least, seven of nine SIV-infected (group 1) and five of six non-SIV-infected macaques (group 2). These findings collectively suggest a strong link between the occurrence of diarrhea and constitutive activation of STAT3. Curiously, constitutively active p-STAT3 proteins were also evident in the jejunum of group 2 animals despite low IL-6 mRNA levels (Figures 1B and 2B and Table 6). This suggests that other cytokines that use the JAK-STAT3 pathway for signaling such as interferon-γ, IL-27, IL-10,38 IL-7, IL-15,51 and chemokines such as RANTES and MIP-1α52 may also be elevated. Their contributions have not been investigated in this study and remain to be determined in the future. Constitutively active STAT3 has also been shown to be an essential mediator of inflammatory bowel disease and various other manifestations of intestinal inflammation in humans.53,54
We next examined which cell types were expressing activated STAT3 using immunohistochemistry and confocal microscopy with anti-phospho-STAT3 antibodies. p-STAT3 expression was restricted to the mononuclear cell population present in the lamina propria. Using cell-specific surface markers such as CD3 and CD68, we found both lymphocytes (CD3) and macrophages (CD68) to be expressing p-STAT3. In the non-SIV-infected macaque with diarrhea, the expression of p-STAT3 was equally distributed between macrophages and lymphocytes (Figure 3, B and E). However, in SIV-infected macaques with moderate (H405) and severe (AT56) CD4+ T-cell depletion, p-STAT3 expression was detectable mostly in macrophages and rarely in CD3+ lymphocytes (Figure 3, A and D; and Supplementary Figure 2, A and B, see http://ajp.amjpathol.org). Interestingly, in an SIV-infected macaque (DI28) with minimal CD4+ T-cell depletion (Table 1) several T cells expressing CD3 were also found to express p-STAT3 (Supplementary Figure 2C, see http://ajp.amjpathol.org) raising the possibility that these may be CD4+ T cells. Further, in both group 1 and 2 animals, we also detected other mononuclear cell populations in the lamina propria resembling lymphocytes that were positive for p-STAT3 but were negative for both CD3 and CD68. In control animals, all p-STAT3 cells were CD3−CD68−. The immunophenotype of these cells remains to be determined. Based on our detection methods, it seems that the constitutive activation of STAT3 is a hallmark of inflammation in the GI tract of SIV-infected macaques and is predominantly expressed by infiltrating macrophages.
Because SOCS-3 is a STAT3-induced gene and also a negative regulator of the JAK-STAT3 pathway, we next investigated the expression pattern of SOCS-3 in both colon and jejunum of all macaques. In the present study, the mRNA expression for SOCS-3 was several fold higher in the colon of SIV-infected and non-SIV-infected macaques with diarrhea than in controls. No elevation in SOCS-3 in the jejunum of group 2 animals compared to controls was observed. However, SOCS-3 in the jejunum of SIV-infected macaques with diarrhea was significantly elevated compared to controls and strongly correlated with the severity of histopathological lesions. This observation agrees with earlier reports in which SOCS-3 mRNA was found to be expressed in the colon of dextran sulfate sodium-treated mice (a model for inflammatory bowel disease in humans) and colonic biopsies from humans with Crohn’s disease or ulcerative colitis patients.55 These findings also suggest that despite high SOCS-3 expression, STAT3 remains active and may not only contribute to the development of inflammation but also delay the beginning of the healing process. It has been suggested that even though SOCS-3 is induced rapidly in response to cytokine signaling, accelerated degradation effectively decreases its half-life and ability to inhibit STAT3 activation.56 Irrespective of the exact mechanism, persistent elevations of p-STAT3 would maintain immune activation in the GI tract, which would be conducive to HIV/SIV replication and disease progression.
In summary, we detected constitutive STAT3 activation in the intestinal tract of SIV-infected and non-SIV-infected macaques with diarrhea. In both groups, SOCS-3 mRNA expression was increased several fold in colon compared to the control group, but in the jejunum, only group 1 animals showed elevation of SOCS-3 compared to controls. These findings suggest that the negative regulation of the IL-6-STAT3 pathway may be dysfunctional in the colon of both groups 1 and 2 but only in the jejunum of group 1. Although IL-6 signaling is important for protecting the gut from infectious agents, its dysregulation can have untoward long-term adverse sequelae in the pathogenesis of GI dysfunction in AIDS patients. In the future, a time-course study using SIV-infected macaques is needed to determine at what point during disease progression the JAK-STAT3 pathway becomes dysregulated. Further, because all group 1 macaques with high IL-6 and constitutive STAT3 expression had high mucosal viral loads, we are also in the process of investigating if the activation of C/EBPβ is part of the molecular mechanism by which IL-6 induces viral replication in GI tract lymphocytes and macrophages.57,58,59 Elucidating the underlying molecular mechanism(s) is critical because macrophages are an important cell type that serve as HIV/SIV reservoirs in chronic HIV-infected patients, especially, after CD4+ T cells have been depleted early in HIV/SIV infection.60 The identification and characterization of key downstream signaling molecules involved in regulating HIV/SIV replication could lead to the development of more attractive therapeutic targets in the future. Finally, our studies also indicate that the rhesus macaque could serve as a good model for inflammatory bowel disease, a chronic inflammatory condition in which activated STAT3 is a critical mediator.53
Supplementary Material
Acknowledgments
We thank Dr. Ronald S. Veazey for providing us with biopsy samples from uninfected healthy macaques and for helpful discussions; Ms. Robin Rodriguez for assistance with the images; Ms. Terri Rasmussen, Lisa Bowers, and Tamika Bridges for technical assistance; and Drs. Preston Marx and Cristian Apetrei, Tulane National Primate Research Center, Division of Microbiology, for their valuable assistance in the study.
Footnotes
Address reprint requests to Andrew A. Lackner, D.V.M., Ph.D., Tulane National Primate Research Center, 18703 Three Rivers Rd., Covington, LA 70433. E-mail: alackner@tulane.edu.
Supported by the National Institutes of Health (grants DK50550, RR00164, RR19607, and AI065325).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- Kewenig S, Schneider T, Hohloch K, Lampe-Dreyer K, Ullrich R, Stolte N, Stahl-Hennig C, Kaup FJ, Stallmach A, Zeitz M. Rapid mucosal CD4(+) T-cell depletion and enteropathy in simian immunodeficiency virus-infected rhesus macaques. Gastroenterology. 1999;116:1115–1123. doi: 10.1016/s0016-5085(99)70014-4. [DOI] [PubMed] [Google Scholar]
- Kotler DP. HIV infection and the gastrointestinal tract. AIDS. 2005;19:107–117. doi: 10.1097/00002030-200501280-00002. [DOI] [PubMed] [Google Scholar]
- Wilcox CM. Chronic unexplained diarrhea in AIDS: approach to diagnosis and management. AIDS Patient Care STDS. 1997;11:13–17. doi: 10.1089/apc.1997.11.13. [DOI] [PubMed] [Google Scholar]
- Oldfield EC., III Evaluation of chronic diarrhea in patients with human immunodeficiency virus infection. Rev Gastroenterol Disord. 2002;2:176–188. [PubMed] [Google Scholar]
- Miao YM, Gazzard BG. Management of protozoal diarrhoea in HIV disease. HIV Med. 2000;1:194–199. doi: 10.1046/j.1468-1293.2000.00028.x. [DOI] [PubMed] [Google Scholar]
- Wilcox CM. Gastrointestinal manifestations of AIDS. Nutr Clin Pract. 2004;19:356–364. doi: 10.1177/0115426504019004356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino A, Bruzzese E, De Marco G, Buccigrossi V. Management of gastrointestinal disorders in children with HIV infection. Paediatr Drugs. 2004;6:347–362. doi: 10.2165/00148581-200406060-00003. [DOI] [PubMed] [Google Scholar]
- Sestak K. Chronic diarrhea and AIDS: insights into studies with non-human primates. Curr HIV Res. 2005;3:199–205. doi: 10.2174/1570162054368084. [DOI] [PubMed] [Google Scholar]
- Heise C, Vogel P, Miller CJ, Halsted CH, Dandekar S. Simian immunodeficiency virus infection of the gastrointestinal tract of rhesus macaques. Functional, pathological, and morphological changes. Am J Pathol. 1993;142:1759–1771. [PMC free article] [PubMed] [Google Scholar]
- Heise C, Miller CJ, Lackner A, Dandekar S. Primary acute simian immunodeficiency virus infection of intestinal lymphoid tissue is associated with gastrointestinal dysfunction. J Infect Dis. 1994;169:1116–1120. doi: 10.1093/infdis/169.5.1116. [DOI] [PubMed] [Google Scholar]
- Lackner AA, Vogel P, Ramos RA, Kluge JD, Marthas M. Early events in tissues during infection with pathogenic (SIVmac239) and nonpathogenic (SIVmac1A11) molecular clones of simian immunodeficiency virus. Am J Pathol. 1994;145:428–439. [PMC free article] [PubMed] [Google Scholar]
- Sasseville VG, Du Z, Chalifoux LV, Pauley DR, Young HL, Sehgal PK, Desrosiers RC, Lackner AA. Induction of lymphocyte proliferation and severe gastrointestinal disease in macaques by a nef gene variant SIVmac239. Am J Pathol. 1996;149:163–176. [PMC free article] [PubMed] [Google Scholar]
- Kuhn EM, Matz-Rensing K, Stahl-Hennig C, Makoschey B, Hunsmann G, Kaup FJ. Intestinal manifestations of experimental SIV-infection in rhesus monkeys (Macaca mulatta): a histological and ultrastructural study. Zentralbl Veterinarmed B. 1997;44:501–512. doi: 10.1111/j.1439-0450.1997.tb01001.x. [DOI] [PubMed] [Google Scholar]
- Kaup F, Matz-Rensing K, Kuhn E, Hunerbein P, Stahl-Hennig C, Hunsmann G. Gastrointestinal pathology in rhesus monkeys with experimental SIV infection. Pathobiology. 1998;66:159–164. doi: 10.1159/000028015. [DOI] [PubMed] [Google Scholar]
- Mattapallil JJ, Smit-McBride Z, McChesney M, Dandekar S. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1beta expression and display antiviral cytotoxic activity despite severe CD4(+) T-cell depletion in primary simian immunodeficiency virus infection. J Virol. 1998;72:6421–6429. doi: 10.1128/jvi.72.8.6421-6429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himathongkham S, Halpin NS, Li J, Stout MW, Miller CJ, Luciw PA. Simian-human immunodeficiency virus containing a human immunodeficiency virus type 1 subtype-E envelope gene: persistent infection. CD4(+) T-cell depletion, and mucosal membrane transmission in macaques. J Virol. 2000;74:7851–7860. doi: 10.1128/jvi.74.17.7851-7860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Byrum RA, Nishimura Y, Endo Y, Plishka RJ, Buckler C, Buckler-White A, Miller G, Hirsch VM, Martin MA. Rapid and irreversible CD4+ T-cell depletion induced by the highly pathogenic simian/human immunodeficiency virus SHIV(DH12R) is systemic and synchronous. J Virol. 2002;76:379–391. doi: 10.1128/JVI.76.1.379-391.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- MacDonald TT, Spencer J. Cell-mediated immune injury in the intestine. Gastroenterol Clin North Am. 1992;21:367–386. [PubMed] [Google Scholar]
- Shanahan F. Intestinal lymphoepithelial communication. Adv Exp Med Biol. 1999;473:1–9. doi: 10.1007/978-1-4615-4143-1_1. [DOI] [PubMed] [Google Scholar]
- Puddington L, Olson S, Lefrancois L. Interactions between stem cell factor and c-Kit are required for intestinal immune system homeostasis. Immunity. 1994;1:733–739. doi: 10.1016/s1074-7613(94)80015-4. [DOI] [PubMed] [Google Scholar]
- Phipps RP, Stein SH, Roper RL. A new view of prostaglandin E regulation of the immune response. Immunol Today. 1991;12:349–352. doi: 10.1016/0167-5699(91)90064-Z. [DOI] [PubMed] [Google Scholar]
- Shanahan F. A gut reaction: lymphoepithelial communication in the intestine. Science. 1997;275:1897–1898. doi: 10.1126/science.275.5308.1897. [DOI] [PubMed] [Google Scholar]
- Wang J, Whetsell M, Klein JR. Local hormone networks and intestinal T cell homeostasis. Science. 1997;275:1937–1939. doi: 10.1126/science.275.5308.1937. [DOI] [PubMed] [Google Scholar]
- Batman PA, Miller AR, Forster SM, Harris JR, Pinching AJ, Griffin GE. Jejunal enteropathy associated with human immunodeficiency virus infection: quantitative histology. J Clin Pathol. 1989;42:275–281. doi: 10.1136/jcp.42.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins AG, LaBrooy JT, Stanley DP, Rowland R, Shearman DJ. Quantitative histological study of enteropathy associated with HIV infection. Gut. 1990;31:317–321. doi: 10.1136/gut.31.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler DP, Reka S, Clayton F. Intestinal mucosal inflammation associated with human immunodeficiency virus infection. Dig Dis Sci. 1993;38:1119–1127. doi: 10.1007/BF01295730. [DOI] [PubMed] [Google Scholar]
- McGowan I, Elliott J, Fuerst M, Taing P, Boscardin J, Poles M, Anton P. Increased HIV-1 mucosal replication is associated with generalized mucosal cytokine activation. J Acquir Immune Defic Syndr. 2004;37:1228–1236. doi: 10.1097/01.qai.0000131846.12453.29. [DOI] [PubMed] [Google Scholar]
- Poli G. Laureate ESCI award for excellence in clinical science 1999. Cytokines and the human immunodeficiency virus: from bench to bedside. Eur Soc Clin Invest Eur J Clin Invest. 1999;29:723–732. doi: 10.1046/j.1365-2362.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- Connolly NC, Riddler SA, Rinaldo CR. Proinflammatory cytokines in HIV disease—a review and rationale for new therapeutic approaches. AIDS Rev. 2005;7:168–180. [PubMed] [Google Scholar]
- Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–368. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109:S121–S131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R. Tuning of macrophage responses by Stat3-inducing cytokines: molecular mechanisms and consequences in infection. Immunobiology. 2005;210:63–76. doi: 10.1016/j.imbio.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Ilangumaran S, Ramanathan S, Rottapel R. Regulation of the immune system by SOCS family adaptor proteins. Semin Immunol. 2004;16:351–365. doi: 10.1016/j.smim.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169–1176. doi: 10.1038/ni1012. [DOI] [PubMed] [Google Scholar]
- May P, Schniertshauer U, Gerhartz C, Horn F, Heinrich PC. Signal transducer and activator of transcription STAT3 plays a major role in gp130-mediated acute phase protein gene activation. Acta Biochim Pol. 2003;50:595–601. [PubMed] [Google Scholar]
- Sestak K, Merritt CK, Borda J, Saylor E, Schwamberger SR, Cogswell F, Didier ES, Didier PJ, Plauche G, Bohm RP, Aye PP, Alexa P, Ward RL, Lackner AA. Infectious agent and immune response characteristics of chronic enterocolitis in captive rhesus macaques. Infect Immun. 2003;71:4079–4086. doi: 10.1128/IAI.71.7.4079-4086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh G, Alvarez X, Borda JT, Aye PP, Lackner AA, Sestak K. Visualizing cytokine-secreting cells in situ in the rhesus macaque model of chronic gut inflammation. Clin Diagn Lab Immunol. 2005;12:192–197. doi: 10.1128/CDLI.12.1.192-197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragione FD, Cucciolla V, Criniti V, Indaco S, Borriello A, Zappia V. p21Cip1 gene expression is modulated by Egr1. A novel regulatory mechanism involved in the resveratrol antiproliferative effect. J Biol Chem. 2003;278:23360–23368. doi: 10.1074/jbc.M300771200. [DOI] [PubMed] [Google Scholar]
- Veazey RS, Tham IC, Mansfield KG, DeMaria M, Forand AE, Shvetz DE, Chalifoux LV, Sehgal PK, Lackner AA. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J Virol. 2000;74:57–64. doi: 10.1128/jvi.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Lackner AA. Getting to the guts of HIV pathogenesis. J Exp Med. 2004;200:697–700. doi: 10.1084/jem.20041464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Interleukin 6 and its receptor: ten years later. Int Rev Immunol. 1998;16:249–284. doi: 10.3109/08830189809042997. [DOI] [PubMed] [Google Scholar]
- Snijders F, van Deventer SJ, Bartelsman JF, den Otter P, Jansen J, Mevissen ML, van Gool T, Danner SA, Reiss P. Diarrhoea in HIV-infected patients: no evidence of cytokine-mediated inflammation in jejunal mucosa. AIDS. 1995;9:367–373. [PubMed] [Google Scholar]
- Olsson J, Poles M, Spetz AL, Elliott J, Hultin L, Giorgi J, Andersson J, Anton P. Human immunodeficiency virus type 1 infection is associated with significant mucosal inflammation characterized by increased expression of CCR5, CXCR4, and beta-chemokines. J Infect Dis. 2000;182:1625–1635. doi: 10.1086/317625. [DOI] [PubMed] [Google Scholar]
- Qin JZ, Kamarashev J, Zhang CL, Dummer R, Burg G, Dobbeling U. Constitutive and interleukin-7- and interleukin-15-stimulated DNA binding of STAT and novel factors in cutaneous T cell lymphoma cells. J Invest Dermatol. 2001;117:583–589. doi: 10.1046/j.0022-202x.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- Wong M, Fish EN. RANTES and MIP-1alpha activate stats in T cells. J Biol Chem. 1998;273:309–314. doi: 10.1074/jbc.273.1.309. [DOI] [PubMed] [Google Scholar]
- Musso A, Dentelli P, Carlino A, Chiusa L, Repici A, Sturm A, Fiocchi C, Rizzetto M, Pegoraro L, Sategna-Guidetti C, Brizzi MF. Signal transducers and activators of transcription 3 signaling pathway: an essential mediator of inflammatory bowel disease and other forms of intestinal inflammation. Inflamm Bowel Dis. 2005;11:91–98. doi: 10.1097/00054725-200502000-00001. [DOI] [PubMed] [Google Scholar]
- Mudter J, Weigmann B, Bartsch B, Kiesslich R, Strand D, Galle PR, Lehr HA, Schmidt J, Neurath MF. Activation pattern of signal transducers and activators of transcription (STAT) factors in inflammatory bowel diseases. Am J Gastroenterol. 2005;100:64–72. doi: 10.1111/j.1572-0241.2005.40615.x. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T, Kubo M, Yamashita A, Okabe M, Takeda K, Akira S, Matsumoto S, Toyonaga A, Sata M, Yoshimura A. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001;193:471–481. doi: 10.1084/jem.193.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander WS. Suppressors of cytokine signaling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- Ito H. IL-6 and Crohn’s disease. Curr Drug Targets Inflamm Allergy. 2003;2:125–130. doi: 10.2174/1568010033484296. [DOI] [PubMed] [Google Scholar]
- Nonnemacher MR, Hogan TH, Quiterio S, Wigdahl B, Henderson A, Krebs FC. Identification of binding sites for members of the CCAAT/enhancer binding protein transcription factor family in the simian immunodeficiency virus long terminal repeat. Biomed Pharmacother. 2003;57:34–40. doi: 10.1016/s0753-3322(02)00334-7. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. Interleukin-6: from basic science to medicine—40 years in immunology. Annu Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- Smith PD, Meng G, Salazar-Gonzalez JF, Shaw GM. Macrophage HIV-1 infection and the gastrointestinal tract reservoir. J Leukoc Biol. 2003;74:642–649. doi: 10.1189/jlb.0503219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.