Abstract
Background
Nitric oxide is released by immune, epithelial and endothelial cells, and plays an important part in the pathophysiology of asthma.
Objective
To investigate the association of inducible nitric oxide synthases (iNOS) gene repeat polymorphisms with asthma.
Methods
230 families with asthma (842 individuals) were recruited to identify and establish the genetic association of iNOS repeats with asthma and associated phenotypes. Serum nitric oxide levels in selected individuals were measured and correlated with specific genotypes. Multiple logistic regression analysis was performed to determine the effect of age and sex.
Results
A total of four repeats—a (CCTTT)n promoter repeat, a novel intron 2 (GT)n repeat (BV680047), an intron 4 (GT)n repeat (AFM311ZB1) and an intron 5 (CA)n repeat (D17S1878)—were identified and genotyped. A significant transmission distortion to the probands with asthma was seen for allele 3 of the AFM311ZB1 gene (p = 0.006). This allele was also found to be significantly associated with percentage blood eosinophils (p<0.001) and asthma severity (p = 0.04). Moreover, it was functionally correlated with high serum nitric oxide levels (p = 0.006). Similarly, the promoter repeat was found to be associated with serum total immunoglobulin (Ig)E (p = 0.028). Individuals carrying allele 4 of this repeat have high serum IgE (p<0.001) and nitric oxide levels (p = 0.03).
Conclusion
This is the first study to identify the repeat polymorphisms in the iNOS gene that are associated with severity of asthma and eosinophils. The functional relevance of the associated alleles with serum nitric oxide levels was also shown. Therefore, these results could be valuable in elucidating the role of nitric oxide in asthma pathogenesis.
Asthma is characterised by increased bronchial responsiveness, constriction and mucus hypersecretion in the bronchial walls in response to a variety of direct and indirect stimuli, leading to the symptoms of cough, wheezing and shortness of breath.1,2 The processes underlying increased airway responsiveness are complex and endogeneous mechanisms may exist to protect against these processes.3 Ricciardolo et al4 have reported that airway‐derived nitric oxide may be important in this respect, as it has a potent bronchodilator action by inducing relaxation of airway smooth muscle. Also, in a mouse model of asthma, it was shown that S‐nitrosothiol (SNO), a relatively stable product of nitric oxide and thiol, is a potent endogeneous bronchodilator.5,6 Inhibition of endogeneous nitric oxide was also shown to increase the airway responsiveness and histamine in patients with asthma.7 However, considerable evidence supports the detrimental effect of nitric oxide. The fraction of nitric oxide in the exhaled air is considered to be a marker of asthma; its magnitude is increased in proportion to bronchial wall inflammation or induced‐sputum eosinophilia as well as to airway responsiveness.8,9 Its levels are reduced in a dose‐dependent manner with anti‐inflammatory treatment; thus, its measurement has been suggested for the maintenance doses of inhaled corticosteroids.10,11
Nitric oxide is synthesised from the semi‐essential amino acid l‐arginine by the enzyme nitric oxide synthase (NOS), of which different isoforms have been identified.12 Constitutive isoforms (collectively called cNOS) generate small amounts of bronchoprotective nitric oxide in response to physiological stimuli, whereas an inducible form of NOS (iNOS, also designated as NOS2A) is induced by inflammatory cytokines during inflammation. iNOS is expressed predominantly in inflammatory cells (T cells and macrophages) and epithelial cells.12 In fact, the epithelial iNOS activity is the major determinant of nitric oxide concentration in exhaled breath.13 Nitric oxide produced by iNOS acts on the submucosal glands to cause mucus secretion. It may act on Th1 cells to down regulate interferon γ production, while acting on Th2 cells to up regulate their production of interleukin (IL)4 and IL5.14,15 In turn, IL4 increases immunoglobulin (Ig)E expression, whereas IL5 recruits eosinophils into the airways.1 Nitric oxide produced by iNOS, which needs a larger amount of substrate, also produces epithelial injury by forming peroxynitrite, which causes airway hyper‐responsiveness.15
The gene for iNOS lies in the CC chemokine cluster region, on chromosome 17q11.2–q12, where a linkage with atopy and asthma has been reported.16,17 Genetic polymorphisms at the NOS2A gene were shown to be associated with susceptibility to diseases such as malaria and multiple sclerosis.18,19 However, a limited number of genetic studies have been reported on atopy or asthma. Konno et al20 suggested that the 14‐repeat allele of CCTTT promoter polymorphism that affects promoter activity is inversely associated with atopy, but not with asthma. Gao et al21 found no association of a biallelic repeat with asthma. The effector role of iNOS in asthma pathology and the results of the previous genetic studies prompted us to analyse the association of various repeat polymorphisms of the iNOS gene with asthma and associated phenotypes.
Material and methods
Participants
In a multicentre‐based asthma genetics study programme, families ascertained through a total of 230 probands (mean age 16.5 (standard deviation (SD) 11.5) years) were recruited from various collaborating hospitals of north and northwest India (Indo‐European). The recruited study population involved children and adults with longstanding asthma (age range 3–48 years). The ethics committees of all the participating centres and hospitals approved the study. Written consent was obtained from all the participants or the parents in the case of children. Study design was such that at least one child with asthma along with both parents were recruited. Thus, in our study, more younger children with a median age of 13.0 years with asthma were recruited. Families were further extended whenever other members gave their consent to participate. Thus, 842 individuals were recruited, with an average family size of 3.47 (range 3–10) individuals per family.
Clinical evaluation of asthma and atopy
In the recruited study population, asthma was defined by clinical history and later validated by interview as described previously.22 Probands were diagnosed with asthma by a primary physician, followed by a confirmatory diagnosis by a common group of clinicians at the time of family visits. Probands were subdivided into three groups—with mild (intermittent or persistent), moderate or severe asthma—on the basis of the National Asthma Education and Prevention Program (Expert Panel Report‐2) guidelines (http://www.nhlbi.nih.gov/guidelines/asthma/asthgdln.pdf). Briefly, clinical tests performed to validate the asthma phenotype were as follows: presence of bronchial hyperresponsiveness (defined as the ratio of forced expiratory volume in one second to forced vital capacity <80% at the time of the episode and improvement by bronchodilators; except for the probands with age <6 years (n = 5) for whom the pulmonary function test was not performed); skin prick test or specific serum IgE (in the case of children); total serum IgE and differential cell count associated with a positive response to at least one of the following questions depending on the age of the patient. Have you ever had episodes of breathlessness at rest with wheezing (mainly in the case of children)? Have you ever had an asthma attack at night? Were you ever admitted to the hospital for asthma? Are you taking any treatment for asthma? Sixteen allergens common to all the centres were used for the skin prick test, with both negative and positive controls (house dust mite, Amaranthus spinosus, Brassica campestris, Cynodon dactylon, Parthenium hysterophorus, Proposis juliflora, Ricinus communis, Alternaria tenuis, Aspergillus fumigatus, cockroach male, cockroach female, mosquito, moth, grain dust rice, hay dust and house dust). Specific serum IgE was estimated for six common allergens (house dust mite, cockroach male, mosquito, moth, grain dust rice and house dust) using the method described by Voller et al,23 with slight modifications. Atopy was defined as a dichotomous variable, having a weal reaction equal to or greater than histamine or high specific IgE for at least one allergen. Total serum IgE levels were estimated using ELISA as described.22
Clinical data on the diagnosis of asthma and atopic diseases or other respiratory disorders, their duration, skin problems, types and doses of drugs and history of tobacco smoking were obtained by completing a detailed questionnaire. Details of environmental factors and the geographical region of origin and migration status were also noted. Individuals with a history of active smoking in the past 3 years or with parasitic or helminthic infestation were excluded from the study. The affected status was also noted for all the family members recruited to the study.
Genomic DNA preparation
DNA was isolated from peripheral blood white cells using the modified salting‐out method, and was stored at −20°C until further analysis, as described previously.22
Identification and genotyping of repetitive sequences in and around the NOS2A gene
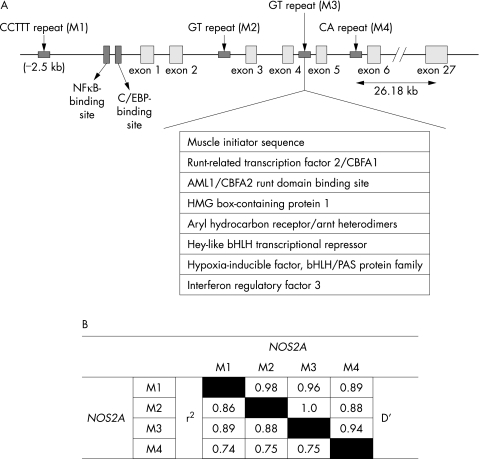
Putative repetitive sequences in and around the NOS2A gene were identified using the RepeatMasker software (http://www.repeatmasker.org/cgi‐bin/WEBRepeatMasker). Four microsatellite repeats—namely, a CCTTT repeat 2.5‐kb upstream from the NOS2A transcription start site (M1), a novel GT repeat (BV680047) in intron 2 (M2), a GT repeat (AFM311ZB1) in intron 4 (M3) and a CA repeat (D17S1878) in intron 5 (M4)—were identified (fig 1) and validated for distribution in our study population by means of a polymerase chain reaction (PCR) using a 6‐FAM‐labelled forward primer and a non‐labelled reverse primer (table 1). Further details can be obtained from the material available online at http://www.thorax.bmjjournals.com/supplemental. The PCR products were separated by electrophoresis through a POP‐4 gel using an ABI 3100 genetic analyser (PE Applied Biosystems, Foster City, California, USA), along with PET‐510 internal size standard. Fragment lengths were determined using the GeneMapper Software V.3.7 (ABI). The number of repeats at each locus was determined by sequencing PCR fragments of individuals (n = 5) being homozygotic for one allele.
Figure 1 (A) Schematic representation of the NOS2A gene. NOS2A is composed of 27 small exons and spans about 43 kb on chromosome 17q11. The region having the promoter and first six exons is expanded to illustrate the position of four repeats genotyped in the study population. Several potential transcription or regulatory factor‐binding sites in intron 4 are shown as predicted by the MatInspector with optimised matrix threshold >85%. (B) Pairwise linkage disequilibrium for all twoway comparisons among the four repeat polymorphisms investigated in the NOS2A gene, with probands with asthma as calculated using EMLD.
Table 1 Primer details for the genotyping of four microsatellite repeats in and around the NOS2A gene.
| Primer name | Number of bases | Primer sequence | Annealing temperature, (°C) | PCR product size (bp) |
|---|---|---|---|---|
| M1_FP | 24 | 5′‐ACCCCTGGAAGCCTACAACTGCAT‐3′ | 62 | 176–221 |
| M1_RP | 24 | 5′‐GCCACTGCACCCTAGCCTGTCTCA‐3′ | ||
| M2_FP | 24 | 5′‐GTGTCATGAAACCAAGAGCCTATT‐3′ | 57 | 215–229 |
| M2_RP | 20 | 5′‐CAGCCCAGCAAGCAACACAT‐3′ | ||
| M3_FP | 21 | 5′‐TTCCCACATGCCCCCAAACCA‐3′ | 57 | 343–339 |
| M3_RP | 22 | 5′‐GCTCCCTGCACCTCTTCTCCAC‐3′ | ||
| M4_FP | 25 | 5′‐AAGGCCATTTTGTTCAGAGTTACCC‐3′ | 62 | 323–341 |
| M4_RP | 26 | 5′‐AGAAAAGAGCTGCCCACGTCATAGTC‐3′ |
NOS2A, inducible nitric oxide synthase; PCR, polymerase chain reaction.
Measurement of serum nitric oxide level
Total serum nitric oxide levels were estimated using a colorimetric assay kit that measures total nitrate, nitrite and SNO as an index of total nitric oxide produced, and hence NOS enzyme activity indirectly (Calbiochem, EMD Biosciences, Darmstadt, Germany). The absorbance was read at 540 nm using the plate reader (SOFT‐MaxPro, Molecular Devices, ELISA Reader, Minnesota, USA). The standard curve was plotted with absorbance against the nitrate concentration. For all statistical analysis, values were log transformed.
Data analysis
Family based association test (http://www.biostat.harvard.edu/∼fbat/fbat.htm) was used to evaluate the association of four markers in NOS2A for the binary traits of asthma or atopy. Biallelic and multiallelic tests were performed using an additive genetic model to identify alleles with evidence for both linkage and association. FBAT analysis extends the methodology of the transmission disequilibrium test to evaluate nuclear families including both affected and unaffected offspring. It conditions on the observed traits and parental genotypes, and where parental data are missing, conditions on the offspring genotype configuration to specify the distribution of a score statistic. The conditional distribution is used to calculate the mean and variance of each family's contribution to a general score statistic. Analysis of variance (ANOVA) was carried out to test the effect of repeat polymorphisms on total serum log10 IgE levels using JMP (SAS Institute, Cary, NC, USA). The alleles significantly associated with the trait in FBAT or with IgE were further evaluated to determine the magnitude and direction of association with serum nitric oxide levels and percentage of blood eosinophil count by ANOVA after making the distribution normal by excluding the extreme outliers using the Shapiro–Wilk test. Association with severity was analysed using a 2×2 table (homozygotic wild type v heterzygote/homozygote mutant) from simple interactive statistical analysis (http://home.clara.net/sisa/twoby2.htm). The influence of the particular polymorphism, age and sex on specific asthma phenotypes was also examined using multivariate logistic regression analysis.
Results
Association of NOS2A microsatellites with asthma
A total of 10, 7, 9 and 11 alleles were observed for the repeats M1, M2, M3 and M4, respectively (table 2).
Table 2 Biallelic results for four markers of iNOS testing null hypotheses of no linkage and no association (family based association test, additive model) in 230 Indian families with probands with asthma.
| Allele | Repeat size | Allele frequency | Informative families (n) | z Score | p Value* |
|---|---|---|---|---|---|
| −2.5 kb CCTTT repeat (M1) | |||||
| 1 (176 bp) | 9 | 0.042 | 32 | 0.926 | 0.355 |
| 2 (181 bp) | 10 | 0.166 | 92 | –0.612 | 0.541 |
| 3 (186 bp) | 11 | 0.162 | 89 | 0.476 | 0.634 |
| 4 (191 bp) | 12 | 0.192 | 108 | –1.273 | 0.203 |
| 5 (196 bp) | 13 | 0.153 | 96 | 0.219 | 0.827 |
| 6 (201 bp) | 14 | 0.132 | 80 | 0.722 | 0.470 |
| 7 (206 bp) | 15 | 0.107 | 61 | 0.663 | 0.508 |
| 8 (211 bp) | 16 | 0.022 | 19 | –0.539 | 0.590 |
| 9 (216 bp) | 17 | 0.012 | 10 | 0.577 | 0.564 |
| 10 (221 bp) | 18 | 0.005 | 2 | – | – |
| Intron 2 GT repeat (M2) | |||||
| 1 (215 bp) | 19 | 0.032 | 24 | –1.095 | 0.273 |
| 2 (217 bp) | 20 | 0.146 | 70 | –0.826 | 0.409 |
| 3 (219 bp) | 21 | 0.522 | 149 | 2.288 | 0.022 |
| 4 (221 bp) | 22 | 0.245 | 104 | –1.422 | 0.155 |
| 5 (223 bp) | 23 | 0.03 | 19 | –0.888 | 0.374 |
| 6 (225 bp) | 24 | 0.014 | 10 | 0.302 | 0.763 |
| 7 (229 bp) | 26 | 0.011 | 2 | – | – |
| Intron 4 GT repeat (M3) | |||||
| 1 (343 bp) | 13 | 0.213 | 89 | –0.926 | 0.354 |
| 2 (345 bp) | 14 | 0.007 | 7 | – | – |
| 3 (347 bp) | 15 | 0.406 | 142 | 2.759 | 0.006 |
| 4 (349 bp) | 16 | 0.166 | 82 | –0.403 | 0.687 |
| 5 (351 bp) | 17 | 0.011 | 9 | – | – |
| 6 (353 bp) | 18 | 0.154 | 88 | –2.001 | 0.045 |
| 7 (355 bp) | 19 | 0.026 | 15 | 0.19 | 0.849 |
| 8 (357 bp) | 20 | 0.016 | 10 | –0.632 | 0.527 |
| 9 (339 bp) | 11 | 0.002 | 1 | – | – |
| Intron 5 CA repeat (M4) | |||||
| 1 (323 bp) | 14 | 0.238 | 65 | 1.73 | 0.08 |
| 2 (325 bp) | 15 | 0.144 | 60 | –1.91 | 0.06 |
| 3 (327 bp) | 16 | 0.207 | 69 | –1.33 | 0.18 |
| 4 (329 bp) | 17 | 0.087 | 28 | –0.82 | 0.41 |
| 5 (331 bp) | 18 | 0.013 | 8 | – | – |
| 6 (333 bp) | 19 | 0.027 | 11 | 2.32 | 0.02 |
| 7 (335 bp) | 20 | 0.046 | 19 | 2.12 | 0.03 |
| 8 (337 bp) | 21 | 0.059 | 30 | –0.26 | 0.79 |
| 9 (339 bp) | 22 | 0.148 | 66 | 0.40 | 0.69 |
| 10 (341 bp) | 23 | 0.029 | 10 | –1.27 | 0.21 |
| 11 (343 bp) | 24 | 0.003 | 1 | — | — |
*p Values are nominal and not corrected for the multiple testing.
Overall, none of the repeats showed a significant association with asthma, when analysed using the multiallelic mode of FBAT (p>0.05). However, we found a significant association of the allele 3 of the M3 repeat with asthma (p = 0.006). Allele 3 was overtransmitted to the probands with asthma as shown by the positive z value of the FBAT analysis. Similarly, allele 3 of the novel GT repeat and allele 6 of the CA repeat showed a positive association, whereas allele 6 of M3 showed a negative association with asthma (p<0.05; table 2). However, these associations were marginally significant. None of the alleles of the promoter CCTTT repeat was found to be associated with asthma in our study. As 86% of our probands with asthma are atopic, further analysis with atopy as such has not been performed.
Allele 3 of M3 in NOS2A and asthma severity
Asthma severity status for the 207 individuals was determined unambiguously. Most (76.3%) of them had mild, 22.2% had moderate and only 1.5% were found to have severe asthma. Allele 3 of M3, which was found to be associated with asthma, was analysed for its association with asthma severity. As only 3 probands with severe asthma were recruited in our study population, they were grouped together with individuals with moderate asthma for association analysis using a 2×2 table. In all, 27% of the individuals carrying allele 3 had moderate or severe asthma, whereas only 12% of those carrying other genotypes fall into this category. Thus, a significant association was observed with asthma severity and allele 3 of M3 (p = 0.02, odds ratio 2.62, 95% confidence interval (CI) 1.03 to 6.6; p = 0.04 after multiple logistic regression).
Associations of the NOS2A repeat polymorphisms with serum total IgE
As increased total serum IgE level is one of the major characteristics of atopy and atopic asthma, and nitric oxide is known to regulate IgE,6 the genetic effects of the NOS2A repeat polymorphisms were tested on this trait. One‐way ANOVA showed a significant association of the promoter CCTTT repeat polymorphism and serum IgE levels (table 3; p = 0.018; p = 0.028 after multiple logistic regression). When mean log10 serum total IgE of individuals with allele 4 was compared with that of individuals with all other alleles pooled together and analysed using ANOVA, we found that the allele 4 (12 repeats) was significantly associated with the high log10 serum total IgE levels (p<0.001). However, none of the other microsatellites (M2, M3 and M4) was found to be associated with IgE in our analysis (table 3).
Table 3 Serum total log10 immunoglobulin (Ig)E levels in the context of alleles of four repeat polymorphisms in NOS2A.
| Allele (repeat size) | Number of alleles | Mean (SD) | Overall p value* |
|---|---|---|---|
| −2.5 kb CCTTT repeat (M1) | |||
| 1 (9) | 24 | 2.87 (0.46) | |
| 2 (10) | 61 | 2.81 (0.51) | |
| 3 (11) | 63 | 2.78 (0.52) | |
| 4 (12) | 76 | 3.09 (0.63) | 0.018 |
| 5 (13) | 55 | 2.87 (0.52) | |
| 6 (14) | 49 | 2.73 (0.53) | |
| 7 (15) | 33 | 2.91 (0.54) | |
| 8 (16) | 10 | 2.85 (0.38) | |
| Intron 2 GT repeat (M2) | |||
| 1 (19) | 8 | 2.74 (0.53) | |
| 2 (20) | 69 | 2.99 (1.00) | |
| 3 (21) | 195 | 2.96 (0.82) | |
| 4 (22) | 86 | 2.96 (0.71) | 0.97 |
| 5 (23) | 12 | 2.91 (0.57) | |
| 6 (24) | 4 | 2.69 (0.66) | |
| Intron 4 GT repeat (M3) | |||
| 1 (13) | 86 | 2.89 (0.47) | |
| 2 (14) | 3 | 2.47 (0.62) | |
| 3 (15) | 174 | 2.96 (0.64) | |
| 4 (16) | 62 | 2.77 (0.53) | |
| 5 (17) | 2 | 2.72 (0.11) | 0.29 |
| 6 (18) | 42 | 2.94 (0.64) | |
| 7 (19) | 11 | 3.03 (0.57) | |
| 8 (20) | 4 | 3.13 (0.15) | |
| Intron 5 CA repeat (M4) | |||
| 1 (14) | 53 | 2.86 (0.56) | |
| 2 (15) | 34 | 2.91 (0.52) | |
| 3 (16) | 80 | 2.88 (0.52) | |
| 4 (17) | 41 | 2.95 (0.45) | |
| 5 (18) | 10 | 2.78 (0.39) | |
| 6 (19) | 22 | 2.83 (0.56) | 0.76 |
| 7 (20) | 25 | 2.89 (0.52) | |
| 8 (21) | 42 | 2.76 (0.51) | |
| 9 (22) | 68 | 2.82 (0.50) | |
| 10 (23) | 4 | 3.18 (0.33 | |
*The p values are nominal and not corrected for the multiple testing.
Association of the NOS2A repeat polymorphisms with peripheral blood eosinophils (%)
We checked for the association of allele 3 of M3 and allele 4 of the M1 with percent blood eosinophil counts. A significant association was observed with allele 3 of M3 (F = 8.64, df = 2, p<0.001); homozygotic individuals had the highest percentage of eosinophils, followed by heterozygotic individuals and then the individuals with other genotypes (table 4). Negligible effects of age and sex were observed when regressed with these two parameters (p<0.001). However, no significant association was observed for the allele 4 of M1 (F = 2.002, df = 2, p = 0.14; table 4).
Table 4 Distribution of the percent peripheral blood eosinophil counts and serum total log10 nitric oxide levels among the individuals with genotypes of allele 3 versus other alleles of M3 and among the individuals with genotypes of allele 4 versus other alleles of M1.
| Percent peripheral blood eosinophil counts | Serum log10 nitric oxide levels | ||||
|---|---|---|---|---|---|
| Number | Mean (SD) | Number | Mean (SD) | ||
| M3 genotype | |||||
| Allele 3/allele 3 | 25 | 8.48 (3.78) | 20 | 1.15 (0.13) | |
| Allele 3/other | 59 | 8.16 (2.45) | 70 | 0.98 (0.20) | |
| Other/other | 31 | 5.74 (2.91) | 52 | 0.90 (0.28) | |
| M1 genotype | |||||
| Allele 4/allele 4 | 3 | 10.7 (1.37) | 8 | 1.16 (0.10) | |
| Allele 4/other | 50 | 7.06 (3.32) | 49 | 1.03 (0.18) | |
| Other/other | 78 | 8.05 (4.07) | 84 | 0.95 (0.24) | |
Functional correlation of the NOS2A repeats with serum levels
Nitric oxide levels were measured in the sera of 241 related and unrelated individuals. Excluding the related individuals, data from 150 unrelated individuals were used for the association analysis of the NOS2A alleles showing association with asthma/IgE. We observed a significant association with allele 3 of M3 (F = 3.28, df = 2, p = 0.04). Also, after excluding the extreme outliers (n = 8) for normalisation of nitric oxide levels using the Shapiro–Wilk test, the association was found to be more significant (F = 8.36, df = 2, p<0.001; p = 0.006 after multiple logistic regression). Accordingly, individuals homozygotic for allele 3 were found to have high levels of nitric oxide in their sera (table 4). Similarly, a significant association was observed for allele 4 of M1 (F = 3.11, df = 2, p = 0.047; F = 5.26, df = 2, p = 0.006 after excluding the outliers (n = 9) using the Shapiro–Wilk test; and p = 0.03 after multiple logistic regression; table 4). The earlier reported 14 repeat of M1 (allele 6) was also found to be marginally associated with serum nitric oxide levels (p = 0.07).24
Discussion
Nitric oxide, a relatively stable free radical, is increased in exhaled air and plasma of people with asthma compared with that in healthy individuals, and is increasingly implicated in the pathogenesis of inflammation in asthma.8,25 Nitric oxide produced in the lungs is an important regulator of airway events, including modifying airway tone, regulating pulmonary vascular tone, stimulating mucin secretion, modulating mucociliary clearance through effects on ciliary beat frequency, and immune surveillance including tumoricidal and bactericidal effects.26 Studies of NOS in asthma have been focused on iNOS as iNOS was shown to be up regulated in patients with asthma and is believed to represent the major source of nitric oxide in the lungs.27 NOS2A (iNOS) has been identified as a calcium‐independent isoform, which was detected in the brain, lungs, and liver of rats after endotoxin treatment.28 The transcriptional activation of iNOS in these cells is regulated by endogeneous mediators (such as chemokines and cytokines) as well as exogenous factors including allergens and environmental pollutants.29
To elucidate the role of iNOS in asthma pathogenesis, we have identified and genotyped four repeats including a novel (GT)n repeat in intron 2 of NOS2A. A significant transmission distortion to the probands with asthma was seen for allele 3 of the (GT)n repeat in intron 4. As the allele at each locus is not fully independent because of linkage disequilibrium across the gene (pairwise r2>0.74 among the repeats; fig 1), and the end points (asthma, IgE, eosinophil counts, asthma severity and nitric oxide levels) used are clearly not independent, no corrections for the multiple testing have been applied in our analysis. Here, we have undertaken a family‐based approach to analyse the association of polymorphic repeats with asthma, which is more robust than the classic population‐based studies. Although population‐based studies have more power, cryptic population stratification can produce false‐positive results. On the other hand, asthma being a complex genetic disorder, owing to small effects of individual genes, a family‐based study may fail to detect significant association even when it exists, because of the power constraints.30 Camp31 suggested that at least 200 informative triads (patients and parents) are required for a completely recessive disorder with a population prevalence rate of 10% and a relative risk (RR) of 0.011. As RR for asthma is >0.01, in order to study 8–10 alleles per locus, the number of families recruited by us (n = 230) would be enough to impart sufficient power (at least 80%) for statistical analysis. Moreover, using the population based association test, the power of our study to detect the association at α = 0.05 for an allele with a frequency of 5% was found to be 82.5%. In addition to FBAT, we have also confirmed our results using other statistical tests such as the pedigree disequilibrium test (data not shown). Moreover, while analysing our data with respect to the other asthma‐associated phenotypes (serum IgE, blood eosinophil counts or serum nitric oxide levels), even after high drop‐outs (small sample size), the power of the statistical analysis was >80% as calculated using the JMP software. Although the samples in our study population were collected from various centres, the samples belong to a homogeneous population of Indo‐European origin. Thus, it is very unlikely that our results are due to stratification or an inherent statistical bias.
Although the association of allele 3 of M3 is novel and interesting, the causal relationship with asthma remains to be elucidated. Various groups have shown that variable length intronic (GT)n repeats can regulate expression or the splicing pattern of a gene.32,33 When analysed using the transcription factor analysis software MatInspector (http://www.genomatix.de/products/MatInspector), we found the presence of binding sites for various regulatory factors such as hypoxia‐inducible factor and interferon regulatory factor 3 near the associated repeat (fig 1).
Interestingly, allele 3 of this repeat was also found to be significantly associated with percent peripheral blood eosinophils as well as with asthma severity. Our results are imperative as nitric oxide is known to modulate the Th1/Th2 balance by favouring Th2 responses and also IL5 production.14 Functional correlations of allele 3 with high serum nitric oxide levels add further confidence to our results. In our study, we measured the total nitric oxide levels in the sera of the selected individuals, which can be influenced by many endogeneous factors, including the three types of NOS and also the SNOs. Nevertheless, as iNOS is the predominant nitric oxide synthesiser in the induced state, the serum total nitric oxide levels could be an indirect reflection of iNOS activity. To gain an insight into the genetic significance of allele 3 of M3 on tissue‐specific expression of nitric oxide, our results have to be further validated using bronchoalveolar lavage fluid, lung biopsy samples or exhaled air. Nevertheless, the possibility of any other repeat polymorphism present within the gene (remaining 20 introns) or any functional small nuclear polymorphisms in linkage disequilibrium with allele 3 of M3 should not be overlooked.
Previous genetic studies have shown the association of the genetic variants of nNOS and eNOS with asthma.21,34 To the best of our knowledge, the only two studies on iNOS repeats have failed to show any association with asthma.20,21 Also, the repeat showing association in our population has not been studied by other groups. In accordance with the findings of Konno et al,20 we have not found any association of the allele 14 and other alleles of the promoter CCTTT repeat with asthma. However, in our study, allele 4 (12 repeat) of this functional promoter repeat polymorphism was found to be associated with high serum total IgE as well as with serum nitric oxide levels. Cardinale et al35 have shown a significant positive correlation between exhaled nitric oxide and total serum IgE in patients with asthma with allele 4. However, the putative role of allele 4 in the transcriptional regulation of iNOS by binding to regulatory proteins remains to be elucidated in the future.
To summarise, this is the first study in any population identifying the association of the iNOS gene repeat polymorphisms with asthma. The associated allele has been further analysed with asthma severity, blood eosinophil counts and, most importantly, with serum nitric oxide levels. In contrast with earlier observations, where nitric oxide was primarily implicated to protect against atopy, in our study, high nitric oxide levels associated with risk genotypes point towards the pro‐inflammatory role of nitric oxide in patients with established asthma.
Supplementary Material
Acknowledgements
JB wishes to thank CSIR for her fellowship. We thank all participating clinicians: Dr SK Sharma, Dr VK Vijayan, Dr PV Niphadkar and Dr V Kumar and the volunteers for helping in this study. We also thank Mr Rajshekhar Chatterjee, Mr A Kumar and Ms Deepti Maan for their technical assistance.
Abbreviations
ANOVA - analysis of variance
FBAT - family‐based association test
iNOS - inducible nitric oxide synthase
NOS - nitric oxide synthases
PCR - polymerase chain reaction
SNO - S‐nitrosothiol
Footnotes
Funding: The Council of Scientific and Industrial Research (Task Force project‐SMM0006), Government of India, provided financial assistance.
Competing interests: None.
References
- 1.Frieri M. Asthma concepts in the new millennium: update in asthma pathophysiology. Allergy Asthma Proc 20052683–88. [PubMed] [Google Scholar]
- 2.Elias J A, Zhu Z, Chupp G.et al Airway remodeling in asthma. J Clin Invest 19991041001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavord I D, Tattersfield A K. Bronchoprotective role for endogenous prostaglandin E2. Lancet 1996345436–438. [DOI] [PubMed] [Google Scholar]
- 4.Ricciardolo F L, Geppetti P, Mistretta A.et al Randomised double‐blind placebo‐controlled study of the effect of inhibition of nitric oxide synthesis in bradykinin‐induced asthma. Lancet 1996348374–377. [DOI] [PubMed] [Google Scholar]
- 5.Gerard C. Biomedicine. Asthmatics breathe easier when it's SNO‐ing. Science 20053081560–1561. [DOI] [PubMed] [Google Scholar]
- 6.Que L G, Liu L, Yan Y.et al Protection from experimental asthma by an endogenous bronchodilator. Science 20053081618–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor D A, McGrath J L, Orr L M.et al Effect of endogenous nitric oxide inhibition on airway responsiveness to histamine and adenosine‐5′‐monophosphate in asthma. Thorax 199853483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payne D N, Adcock I M, Wilson N M.et al Relationship between exhaled nitric oxide and mucosal eosinophilic inflammation in children with difficult asthma, after treatment with oral prednisolone. Am J Respir Crit Care Med 20011641376–1381. [DOI] [PubMed] [Google Scholar]
- 9.Jatakanon A, Lim S, Kharitonov S A.et al Correlation between exhaled nitric oxide, sputum eosinophils, and methacholine responsiveness in patients with mild asthma. Thorax 19985391–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S L, Kittelson J, Cowan J O.et al The predictive value of exhaled nitric oxide measurements in assessing changes in asthma control. Am J Respir Crit Care Med 2001164738–743. [DOI] [PubMed] [Google Scholar]
- 11.Kharitonov S A, Donnelly L E, Montuschi P.et al Dose‐dependent onset and cessation of action of inhaled budesonide on exhaled nitric oxide and symptoms in mild asthma. Thorax 200257889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dweik R A. Nitric oxide, hypoxia, and superoxide: the good, the bad, and the ugly! Thorax 200560265–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane C, Knight D, Burgess S.et al Epithelial inducible nitric oxide synthase activity is the major determinant of nitric oxide concentration in exhaled breath. Thorax 200459757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y, Karupiah G, Hogan S P.et al Inhibition of allergic airway inflammation in mice lacking nitric oxide synthase 2. J Immunol 1999162445–452. [PubMed] [Google Scholar]
- 15.Duguet A, Iijima H, Eum S Y.et al Eosinophil peroxidase mediates protein nitration in allergic airway inflammation in mice. Am J Respir Crit Care Med 20011641119–1126. [DOI] [PubMed] [Google Scholar]
- 16.The Collaborative Study on the Genetics of Asthma ( C S G A ) A genome‐wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 199715389–392. [DOI] [PubMed] [Google Scholar]
- 17.Dizier M H, Besse‐Schmittler C, Guilloud‐Bataille M.et al Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med 20001621812–1818. [DOI] [PubMed] [Google Scholar]
- 18.Burgner D, Usen S, Rockett K.et al Nucleotide and haplotypic diversity of the NOS2A promoter region and its relationship to cerebral malaria. Hum Genet 2003112379–386. [DOI] [PubMed] [Google Scholar]
- 19.Barcellos L F, Begovich A B, Reynolds R L.et al Linkage and association with the NOS2A locus on chromosome 17q11 in multiple sclerosis. Ann Neurol 200455793–800. [DOI] [PubMed] [Google Scholar]
- 20.Konno S, Hizawa N, Yamaguchi E.et al (CCTTT)n repeat polymorphism in the NOS2 gene promoter is associated with atopy. J Allergy Clin Immunol 2001108810–814. [DOI] [PubMed] [Google Scholar]
- 21.Gao P S, Kawada H, Kasamatsu T.et al Variants of NOS1, NOS2, and NOS3 Genes in asthmatics. Biochem Biophys Res Commun 2000267761–763. [DOI] [PubMed] [Google Scholar]
- 22.Nagpal K, Sharma S, Rao C B.et al TGFβ1 haplotypes and asthma in Indian populations. J Allergy Clin Immunol 2005115527–533. [DOI] [PubMed] [Google Scholar]
- 23.Voller A, Bidwell D, Bartlett A. A serological study on human Trypanosoma rhodesiense infections using a micro‐scale enzyme linked immunosorbent assay. Tropenmed Parasitol 197526247–251. [PubMed] [Google Scholar]
- 24.Warpeha K M, Xu W, Liu L.et al Genotyping and functional analysis of a polymorphic (CCTTT)n repeat of NOS2A in diabetic retinopathy. FASEB J 1999131825–1832. [DOI] [PubMed] [Google Scholar]
- 25.Kocyigit A, Zeyrek D, Keles H.et al Relationship among manganese, arginase, and nitric oxide in childhood asthma. Biol Trace Elem Res 200410211–18. [DOI] [PubMed] [Google Scholar]
- 26.Nathan C, Xie Q W. Nitric oxide synthases: roles, tolls, and controls. Cell 199478915–918. [DOI] [PubMed] [Google Scholar]
- 27.Yates D H, Kharitonov S A, Thomas P S.et al Endogenous nitric oxide is decreased in asthmatic patients by an inhibitor of inducible nitric oxide synthase Am J Respir Crit Care Med1996154247–250. [DOI] [PubMed] [Google Scholar]
- 28.Knowles R G, Merrett M, Salter M.et al Differential induction of brain, lung and liver nitric oxide synthase by endotoxin in rat. Biochem J 1990270833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricciardolo F L, Sterk P J, Gaston B.et al Nitric oxide in health and disease of the respiratory system. Physiol Rev 200484731–765. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Gastwirth J L, Gail M H. Power and related statistical properties of conditional likelihood score tests for association studies in nuclear families with parental genotypes. Ann Hum Genet 200569296–314. [DOI] [PubMed] [Google Scholar]
- 31.Camp N J. Genomewide transmission/disequilibrium testing—consideration of the genotypic relative risks at disease loci. Am J Hum Genet 1997611424–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa‐Mallena, Kelada S N, Costa L G.et al Characterization of the in vitro transcriptional activity of polymorphic alleles of the human monoamine oxidase‐B gene. Neurosci Lett 2005383171–175. [DOI] [PubMed] [Google Scholar]
- 33.Gabellini N. A polymorphic GT repeat from the human cardiac Na+Ca2+ exchanger intron 2 activates splicing. Eur J Biochem 20012681076–1083. [DOI] [PubMed] [Google Scholar]
- 34.Leung T F, Liu E K, Tang N L.et al Nitric oxide synthase polymorphisms and asthma phenotypes in Chinese children. Clin Exp Allergy 2005351288–1294. [DOI] [PubMed] [Google Scholar]
- 35.Cardinale F, Benedictis F M, Muggeo V.et al Exhaled nitric oxide, total serum IgE and allergic sensitization in childhood asthma and allergic rhinitis. Pediatr Allergy Immunol 200516236–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.