Abstract
Prokaryotic chromosomes and plasmids encode partitioning systems that are required for DNA segregation at cell division. The systems are thought to be functionally analogous to eukaryotic centromeres and to play a general role in DNA segregation. The parA system of plasmid R1 encodes two proteins ParM and ParR, and a cis-acting centromere-like site denoted parC. The ParR protein binds to parC in vivo and in vitro. The ParM protein is an ATPase that interacts with ParR specifically bound to parC. Using electron microscopy, we show here that parC mediates efficient pairing of plasmid molecules. The pairing requires binding of ParR to parC and is stimulated by the ParM ATPase. The ParM mediated stimulation of plasmid pairing is dependent on ATP hydrolysis by ParM. Using a ligation kinetics assay, we find that ParR stimulates ligation of parC-containing DNA fragments. The rate-of-ligation was increased by wild type ParM protein but not by mutant ParM protein deficient in the ATPase activity. Thus, two independent assays show that parC mediates pairing of plasmid molecules in vitro. These results are consistent with the proposal that replicon pairing is part of the mechanism of DNA segregation in prokaryotes.
Keywords: partitioning/centromere
Efficient DNA segregation is required for stable inheritance of genetic material to the progeny cells at cell division. In eukaryotes many components of the segregation apparatus have been identified and characterized (1–5). In contrast, the molecular apparatus securing DNA segregation in prokaryotes is not as well understood. Several genes involved in DNA segregation have been identified, but the molecular mechanisms by which the genes function are not known. The prokaryotic systems are more difficult to study due to the lack of visible structures and in vitro systems that can imitate parts of the DNA segregation process (6, 7). The best examined determinants of DNA segregation in prokaryotes are partitioning systems of low-copy-number plasmids, such as par of plasmid P1 (8), sop of F (9), and parA of R1 (10). These systems all consist of a cis-acting site and two genes that encode proteins required for the partitioning process. Here we use the parA partitioning system of plasmid R1 to examine the molecular mechanism of DNA segregation. The parA system is composed of a cis-acting centromere-like site, parC, and two genes encoding the trans-acting proteins, ParM and ParR (11). The 160-bp parC site is located immediately upstream of the two genes and contains the parA promoter flanked by two sets of five direct repeats (iterons), see Fig. 1. The ParR protein binds specifically to the ten iterons in parC (12–13). The presence of all 10 iterons is required for full plasmid stabilization, for incompatibility toward other parA carrying plasmids and for autoregulation of the parA promoter (11–13). ParM possesses an ATPase activity that is required for plasmid partitioning. The ParM protein interacts with ParR bound to parC and is probably present in the protein-DNA complex formed at parC (14). Similar functions have been described for the ParA and ParB partitioning proteins of P1 and the SopA and SopB proteins of F (15–18). In the P1 par and F sop systems, the centromere-like sites are located downstream of the genes encoding the partitioning proteins (8–9).
Figure 1.
Schematic overview of the parA partitioning system of plasmid R1. □, parM gene; ┌, parR gene; and |→, parA promoter. The full-length parC region and the truncated parC sites used in this work are shown. Iterons are indicated with → and the −35 and −10 regions of the parA promoter are indicated with —.
Recently, chromosomal genes that are homologous to plasmid partitioning systems have been identified in a large number of bacteria and shown to be involved in chromosome segregation in Bacillus subtilis (soj and spo0J) and Caulobacter crescentus (parA and parB) (19–21). The partitioning proteins Spo0J of B. subtilis, ParA and ParB of C. crescentus, as well as the origin-proximal regions of the chromosomes of B. subtilis and Escherichia coli localize toward the poles of the cell (21–28). In B. subtilis, the Spo0J protein is involved in maintaining the bipolar localization of the origin-proximal region. The P1 and F plasmids and the SopB partitioning protein of F are positioned at midcell or quarter positions in the cell (26, 29, 30). Rapid movement of newly replicated chromosomal origins or plasmids has been observed, indicating that the replicons are actively separated (31, 32).
Current models describing DNA segregation by plasmid and chromosomal partitioning systems suggest that one or both of the partitioning proteins bind to the cis-acting centromere-like site and promote pairing of replicon copies via protein-protein interactions (12, 28, 33). Here, we demonstrate that the cis-acting parC site of plasmid R1 mediates efficient pairing of plasmid molecules. Replicon pairing requires the presence of the ParR partitioning protein and is stimulated by the second plasmid-encoded partitioning protein ParM.
MATERIALS AND METHODS
Plasmids.
Plasmid pMD330 (11) contains as the only R1 derived sequence the minimal parC region cloned into pUC19 (34). Plasmid pAB1922 contains parC22, in which the spacer region of 39 bp between the two sets of direct repeats has been deleted (13) and pMD333 contains parC33, in which the five downstream repeats of parC have been deleted (11).
Electron Microscopy Analysis.
Supercoiled or SspI digested pMD330 DNA (40–500 ng) was incubated with ParR (25–500 ng), ParM (25–500 ng), ParM D170E (100–250 ng), or E. coli RNA polymerase (40–250 ng) for 15 min at 37°C in 20 μl of 30 mM triethanolamine⋅HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, and, when appropriate, 1 mM ATP, ADP, adenylyl-imidophosphat, or adenosine-5′-O-(3-thiotriphosphate). The complexes were subsequently fixed with 0.2% glutaraldehyde for 15 min at 37°C and the glutaraldehyde was removed by using Micro Bio-Spin 30 gel filtration columns (Bio-Rad). After cleavage of the DNA with SspI, the gel filtration step was repeated. Adsorption to mica, rotational shadowing with platinum, and subsequent evaluation was carried out as described (35). In experiments with linear DNA the complexes were adsorbed to mica directly after fixation. In the experiments where the contour-length of ParR bound fragments were measured, the DNA was cut with PvuII instead of SspI to yield small parC-containing fragments.
Ligation Kinetics Assay.
Blunt-ended radioactively labeled DNA fragments (10 ng) were incubated with or without ParR (100 ng), ParM (200 ng), or ParM D170E (200 ng) for 15 min at 30°C in 20 μl of 10 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 12% PEG 6000, 50 μg/ml BSA, and 15 μg/ml sonnicated salmon sperm DNA. Then 2 units T4 DNA-ligase were added and the reactions were incubated 15 min at 30°C. The reactions were terminated by addition of 20 μl of 10 mM EDTA followed by phenol/chloroform extraction and the DNA was loaded onto a 2% MetaPhor agarose gel (FMC Bioproducts). The radioactivity was quantified by using a Storm 840 PhosphorImager (Molecular Dynamics). The quantitative data reported are averages of at least three independent determinations. The radiolabeled blunt-ended fragments were made by digesting uniformly labeled PCR products with HincII. For the parC containing fragment the primers M13revers: AACAGCTATGACCATG, parA172B: CCCCGTTAACACCAACATTTATAAAACTC and the template pDD19 (11) were used and for the control fragment the primers M13revers: AACAGCTATGACCATG, pUC-control: CCCCGTCGACGAGTGCACCATATGCGGT and the template pUC19 were used in PCR reactions containing α-32P-dCTP. In the experiments where the orientation of the ligated parC fragments was examined, 15% PEG 6000 was used when the fragment was ligated without ParR and ParM. Half of the ligation reactions were digested with BamHI and the DNA was loaded on a 3% MetaPhor agarose gel.
RESULTS
ParR Binds Specifically to the Centromere-Like parC Site.
We investigated the protein-DNA complexes formed between ParR and parC by using electron microscopy (Fig. 2). Depending on the actual experiment, 43% to 76% of linear parC containing DNA molecules had ParR bound specifically to a single site in the plasmid. Measurements of the binding-position confirmed that ParR interacts specifically with the parC site (Fig. 3). Control DNA (pUC19) that does not contain parC did not exhibit such binding. When ParR was bound to supercoiled DNA and the DNA subsequently was linearized (Fig. 2A), the protein-DNA complexes appeared more compact than when ParR was bound to prelinearized DNA (Fig. 2B). Contour-length-measurements of the protein-bound DNA molecules showed that the length of the DNA was reduced by ≈130 bp compared with that of free DNA molecules (Table 1). The shortening was apparent only when ParR was bound to supercoiled DNA, whereas binding of ParR to linear parC DNA or to supercoiled DNA containing truncated parC derivatives (parC22 and parC33, see Fig. 1) did not shorten the DNA (Table 1). These results are consistent with the proposal that the parC DNA is wrapped around a core of ParR molecules only on supercoiled DNA and only when the entire parC region is present.
Figure 2.
Electron micrographs of protein-DNA complexes formed at the parC centromere-like site of plasmid R1 showing ParR binding (A and B), specific pairing of two DNA molecules (C–E) or a higher-order-complex containing multiple DNA molecules (F). ParR and/or ParM were incubated with the parC containing plasmid pMD330 and prepared for electron microscopy. The plasmid was linearized at a unique SspI site to generate a fragment in which the parC region is located asymmetrically. The linearization with SspI was performed either before or after addition of proteins as described below. Bar = 1 kb. (A) ParR bound to parC prepared by incubation of supercoiled pMD330 plasmid DNA with ParR and linearization after binding. (B) ParR bound to parC prepared by incubation of ParR with prelinearized pMD330 DNA. (C) Paired pMD330 plasmid molecules prepared by incubating supercoiled pMD330 with ParR followed by linearization. (D) Paired plasmid molecules prepared by incubating supercoiled pMD330 DNA with ParR, ParM, and ATP. (E) Paired pMD330 molecules formed with prelinearized DNA in the presence of ParR, ParM, and ATP. (F) Typical higher-order-complex where three or more parC DNA molecules are aggregated by ParR protein.
Figure 3.
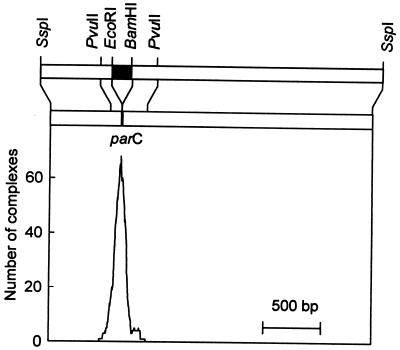
Measurement of the position of ParR binding. ParR was bound to supercoiled pMD330 DNA, the DNA was linearized with SspI, prepared for electron microscopy and the distances from the DNA ends to the ParR binding position were measured. The histogram represents the measurement of binding positions of ParR to 270 molecules. Shown above the graph is a schematic drawing of the DNA fragment used for electron microscopy. The parC region is shown as ■ and the restriction sites relevant to this work are indicated. The upper bar represents the free fragment and the lower bar the ParR-bound fragment, which appeared shortened by 130 bp as compared with the free fragment (Table 1). The shortening of the ParR-bound parC fragment is consistent with DNA wrapping (see Materials and Methods). Bar = 500 bp.
Table 1.
Contour length of ParR bound parC-containing fragments
| parC DNA used | Fragment size | Measured length of ParR-bound fragment | Molecules measured, n |
|---|---|---|---|
| Supercoiled parC | 466 | 335 ± 38 | 266 |
| Linear parC | 466 | 456 ± 22 | 287 |
| Supercoiled parC22 | 439 | 419 ± 26 | 170 |
| Supercoiled parC33 | 390 | 371 ± 25 | 164 |
ParR (100 ng) was bound to 100 ng pMD330 DNA (wt parC), pAB1903 DNA (parC22), or pMD333 DNA (parC33) and prepared for electron microscopy. The contour lengths in bp (±SD) of free and protein-bound DNA fragments were measured and analysed statistically.
Pairing of Plasmid Molecules by ParR.
In addition to ParR bound DNA monomers, we also observed paired structures in which two parC-containing DNA molecules were joined at a protein complex (Fig. 2 C–E). The intersection corresponded to the location of parC. When supercoiled parC DNA was incubated with ParR, 40–45% of the protein-bound DNA molecules were present as paired structures. When linear instead of supercoiled DNA was used, the frequency of pairing was reduced to 6–14% depending on the experimental conditions (Table 2 and Fig. 4). When supercoiled DNA containing truncated parC sites (parC22 or parC33) were used, DNA binding was still efficient but a lower pairing frequency was observed (Table 2). This result indicates that pairing is more efficient between wrapped parC complexes.
Table 2.
Quantification of protein-mediated cohesion of parC DNA molecules
| parC DNA used | Proteins and nucleotides added | % of protein-bound DNA molecules present as
|
Molecules counted, n | |
|---|---|---|---|---|
| Pairs | Higher order complexes | |||
| Supercoiled parC | ParR | 45 | 2.2 | 138 |
| Supercoiled parC | ParR + ParM + ATP | 52 | 2.5 | 202 |
| Supercoiled parC22 | ParR | 23 | 1.5 | 197 |
| Supercoiled parC33 | ParR | 10 | 1.9 | 158 |
| Linear parC | ParR | 5.5 | 0.0 | 181 |
| Linear parC | ParR + ParM + ATP | 12 | 0.0 | 212 |
| Linear parC | RNA polymerase | 0.7 | 0.0 | 289 |
ParR (100 ng), ParM (100 ng), and/or RNA polymerase (125 ng) were bound to 100 ng supercoiled or linearized pMD330 DNA (wt parC), pAB1922 (parC22), or pMD333 DNA (parC33) and prepared for electron microscopy. The frequencies of protein-bound DNA molecules present in paired structures or higher-order-complexes were determined by counting randomly selected fields. Shown are the results from a single, representative experiment.
Figure 4.
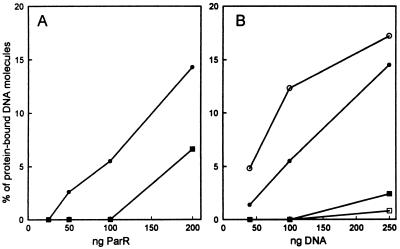
Pairing efficiencies at different concentrations of ParR and DNA measured by electron microscopy. ○ and • indicate % of protein-bound DNA molecules present as paired structures and □ and ■ represent % of protein-bound DNA molecules present as higher-order-complexes. • and ■ indicate the frequencies in the presence of ParR alone and ○ and □ represent the frequencies in the presence of ParR, ParM, and ATP. More than 250 molecules were counted for each data point. In A, 100 ng linearized pMD330 DNA were incubated with the amounts of ParR as indicated. In B, the indicated amounts of linearized pMD330 DNA were incubated with ParR and when relevant ParM and ATP. In B, the amounts of ParR and ParM were titrated for each DNA concentration to optimize DNA binding and pairing.
None or very few higher-order-complexes containing more than two aggregated plasmid molecules (Fig. 2F) were observed by electron microscopy under the conditions used here (Table 2 and Fig. 4). The low abundancy of higher-order-complexes indicates that binary pairing is a specific phenomenon.
E. coli RNA polymerase, which binds to specific sites (i.e., promoters) in the DNA used, was included as a control. RNA polymerase did not mediate significant cohesion of parC containing DNA molecules (Table 2). Similar low frequencies of pairing were observed when other DNA binding proteins not expected to give pairing were used (not shown). Thus the efficient DNA pairing mediated by ParR is not caused by the method of preparation.
ParM Increases the Pairing Frequency in the Presence of ATP.
Only ParR was required for the binary interaction of plasmid molecules, but the pairing-frequency was increased when ParM and ATP also were present (Table 2 and Fig. 4B). Thus, in the presence of ParR, ParM, and ATP up to 64% of the protein-bound DNA molecules were present as paired complexes. The stimulatory effect of ParM was largest under conditions in which ParR alone yielded a modest degree of pairing by itself, i.e., when linear DNA was used or at low ParR concentrations (Table 2 and Fig. 4). Thus, at low concentrations of ParR, ParM, and ATP stimulated pairing up to 3-fold. The modest stimulation by ParM at conditions where ParR alone gives efficient pairing was expected, because ParR alone formed close to saturating amounts of paired complexes.
The ParM mediated enhancement of pairing required ATP, as the pairing frequency was not stimulated by ParM in the absence of the nucleotide, in the presence of ADP or in the presence of the nonhydrolyzable ATP analogs adenylyl-imidophosphate or adenosine-5′-O-(3-thiotriphosphate) (Table 3). Additionally, the ParM D170E mutant protein, which exhibits reduced ATPase activity (14), did not increase pairing even in the presence of ATP. Thus, it appears that ATP hydrolysis by ParM is required for the ParM-mediated enhancement of pairing. The strict dependency of ATP for the ParM mediated stimulation of pairing indicates that pairing is a specific phenomenon and that ParM may have a function in plasmid pairing in vivo.
Table 3.
Quantification of the nucleotide dependency of the ParM mediated enhancement of pairing
| Proteins used | Nucleotide used | % of protein-bound DNA molecules present as
|
Molecules counted, n | |
|---|---|---|---|---|
| Pairs | Higher order complexes | |||
| ParR | — | 6.3 | 0.0 | 253 |
| ParR + ParM | — | 4.7 | 0.0 | 253 |
| ParR + ParM | ATP | 11 | 0.0 | 229 |
| ParR + ParM | ADP | 6.3 | 0.0 | 224 |
| ParR + ParM | AMP-PNP | 7.1 | 0.0 | 225 |
| ParR + ParM | ATPγS | 5.7 | 1.2 | 243 |
| ParR + ParM D170E | ATP | 5.6 | 0.0 | 259 |
ParR (100 ng), ParM (100 ng), and/or ParM D170E (100 ng) were bound to 100 ng linearized pMD330 DNA in the presence of the indicated nucleotides and prepared for electron microscopy. AMP-PNP and ATPγS are nonhydrolysable ATP analogs. The frequencies of protein-bound DNA molecules present in paired structures or higher-order-complexes were determined by counting randomly selected fields. Shown are the results from a single, representative experiment. AMP-PNP, adenylyl-imidophosphate; ATPγS, adenosine-5′-O-thiotriphosphate.
Variation of the Concentrations of the Components in the Pairing Reaction.
The highest pairing frequencies were observed at high concentrations of ParR and parC DNA (Fig. 4 A and B). This result most likely reflects the increased rate by which ParR bound parC DNA monomers meet and form paired complexes. Varying the ParM concentration within a 20-fold range had no effect on the pairing frequency (not shown). At very high concentrations of ParR we observed aggregation of protein on the DNA and increased amounts of higher-order-complexes (Fig. 2F and Fig. 4A). Such aggregation could be caused by binding of ParR to the regions next to parC thereby allowing the simultaneous pairing of more than two molecules.
ParR and ParM Stimulates Ligation of parC Containing DNA Fragments.
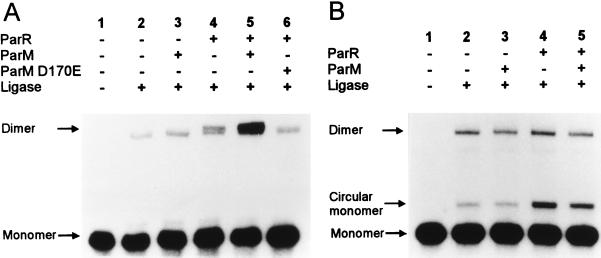
To further assess the ability of ParR and ParM to connect two parC containing DNA fragments, a ligation kinetics assay was performed (Fig. 5A). In this assay, short blunt-ended DNA fragments were incubated with the partitioning proteins and T4 DNA ligase. If the proteins added pair the DNA fragments, the rate of intermolecular ligation increases because of the proximity of the DNA ends (36). When a parC containing DNA fragment was used, ParR increased the amount of ligated products (average, 2.2-fold increase). Addition of both ParR and ParM increased the amount of ligated products further (average 6-fold increase). The ATPase activity of ParM was required for this increase, because addition of ParM D170E did not stimulate ligation (Fig. 5A, lane 6). These results corroborate the conclusions drawn from the experiments using electron microscopy. When a DNA fragment that did not contain parC was used in the ligation kinetics assay, ParR and ParM did not stimulate dimer formation (Fig. 5B). Circularization of the monomer control-fragment was stimulated by ParR. This could be caused by bending of the DNA fragment by nonspecifically bound ParR molecules.
Figure 5.
T4 DNA-ligase mediated multimerization of blunt-end DNA fragments. In A, a 203-bp parC containing fragment with short equally sized arms was used and in B, a 203-bp fragment from pUC19 was used as a nonspecific control DNA. Shown are autoradiograms of agarose gels with the ligation products formed in the presence or absence of ParR, ParM, and the mutated ParM D170E protein as indicated. The dimer band obtained in A consists of two closely spaced bands that correspond to a linear dimer and a circular dimer, respectively.
The Binary ParR-ParM-parC Complex Has a Specific Orientation.
To examine whether the pairing mediated by the Par proteins was orientation-specific, we exploited that the parC DNA fragment used in the ligation kinetics assay contains an asymmetrically located BamHI restriction site. As shown in Fig. 6A, three types of ligated dimer fragments form during ligation. The expected relative frequencies of the three types of dimers when ligation is random are also shown in Fig. 6A. Cleavage with BamHI was used to discriminate between head-to-head inverted dimers and the other two forms of dimers (Fig. 6). The concentrations of the molecular crowding agent PEG in the ligation reactions were adjusted to give equal amounts of multimeric DNA in the presence and in the absence of the partitioning proteins (see Materials and Methods). When the parC-fragment was ligated in the absence of ParR and ParM, purified and restricted with BamHI, 29% of the multimeric DNA was present in the 386-bp band that corresponded to a head-to-head inverted dimer (top fragment in Fig. 6A). This frequency is compatible with random formation of dimers. However, when the parC-fragment was ligated in the presence of ParR and ParM and treated similarly, 94% of the multimeric DNA was present in the head-to-head inverted dimer band (Fig. 6B). This is significantly higher than the expected (25%) and observed (29%) frequencies when ligation was random, thus indicating that ParR and ParM orients the parC DNA in the binary complexes in a nonrandom fashion.
Figure 6.
Orientation-specific ligation of parC fragments. (A) Schematic representation of the possible orientations of ligated dimer parC DNA fragments. The iterons are indicated as triangles and the asymmetrically located BamHI site is indicated with a B. The numbers (in %) are theoretical proportions of the different fragments if ligation is random. (B) DNA fragments containing the parC region were ligated in the absence (lanes 1 and 3) or in the presence (lanes 2 and 4) of ParR and ParM. The reactions were performed such that equal amounts of the ligated multimeric fragments were obtained in lanes 1 and 2 (see Materials and Methods). Lanes 1 and 2 show undigested ligation products. The ligated fragments were treated with BamHI, which cleaves 10 bp from one end of the monomer fragment (lane 3 and 4). The same amount of ligation reaction was loaded in all lanes. The 386-bp fragment in lanes 3 and 4 corresponds to a head-to-head inverted dimer in which two monomer fragments were ligated end to end opposite to the BamHI site (top fragment in A). The position of the circular dimer band, which runs above the linear dimer band, is indicated.
DISCUSSION
The mechanism of DNA segregation in prokaryotes remains elusive. Previously, we proposed that partitioning by the parA system of plasmid R1 involves pairing of plasmid molecules before segregation to the progeny cells (12). Similar pairing models have been proposed for other plasmid and chromosome partitioning systems (28, 29, 33). Using two independent assays, electron microscopy of protein-DNA complexes and a ligation kinetics assay, we obtained results showing that two parC containing DNA molecules are paired by the plasmid-encoded partitioning proteins in vitro. Pairing required only binding of ParR to the centromere-like parC site. The pairing most likely involves protein-protein interactions between ParR molecules bound to the two plasmids. In the presence of ATP, ParM stimulated pairing specifically. The ParM mediated increase of pairing was largest at low concentrations of DNA and ParR (Fig. 4). We have shown previously that ParM interacts specifically with ParR bound to parC (14). Possibly, ATP bound ParM could stimulate pairing by interacting with ParR and thereby stabilizing the paired complex. We believe that the observed replicon-pairing is a specific phenomenon that reflects the in vivo functions of the parA system, because of the following observations: (i) in two independent assays, ParR yielded pairing of parC-containing DNA fragments; (ii) very efficient pairing was observed with supercoiled DNA (up to 64% of the protein-bound DNA molecules were present as paired structures) virtually in the absence of higher-order-complexes; (iii) pairing was specifically increased by the second plasmid encoded partitioning protein ParM in the presence of ATP. This specificity is also inconsistent with pairing being a consequence of nonspecific aggregation; (iv) in the ligation kinetics assay, monomers were ligated to dimers in a nonrandom orientation. This result suggests that the paired protein-DNA complex had a specific structure. Together, our results clearly indicate that the observed plasmid pairing is caused by intrinsic properties of the partitioning proteins and not by nonspecific aggregation. Thus, ParR and ParM mediated association of two parC containing DNA molecules is likely to be a part of the partitioning process in vivo.
When ParR was bound to supercoiled DNA, the protein-DNA complexes were very compact and the contour-lengths of the DNA molecules were shortened by ≈130 bp (Fig. 2 and Table 1). This is consistent with wrapping of the parC DNA around a core of ParR protein. When linear parC DNA or supercoiled DNA containing truncated parC sites were used, shortening of the DNA molecules were not observed and the protein-DNA complexes were less compact. Thus, wrapping (i.e., shortening) presumably requires supercoiled DNA and full-length parC (Table 1). Because pairing was most efficient when supercoiled parC DNA was used, the presumed wrapped complex may have a conformation in which interactions between parC-bound ParR molecules are more efficient. This may also explain the observation that only a full-length parC site is fully active in in vivo partitioning (11, 13). We cannot exclude that the truncated parC sites can form unstable wrapped complexes on supercoiled DNA, that are less efficient in pairing and that cannot be detected by using electron microscopy. Wrapping was previously observed in the sop partitioning system of the F plasmid, where the centromere-like sopC site is wrapped around a core of SopB protein (37–39). The wrapping was evident in vivo, but no wrapping was observed in vitro when linear DNA was used (40).
All known partitioning systems possess an interference phenotype called incompatibility. The phenotype is associated with the centromere-like sites and is manifested as destabilization of a test-plasmid by the centromere-like site present on a second, heterologous plasmid in the same cell line (41). The observed interference with the function of the partitioning system is most readily explained as being a consequence of centromere-mediated pairing of heterologous replicons. Our observations support that mechanism for partitioning associated incompatibility. This suggestion is further supported by the finding that the truncated parC33 site neither gives efficient pairing in vitro (Table 2) nor exerts significant incompatibility toward plasmids stabilized by parA (13).
Information on plasmid pairing in prokaryotes was obtained during the study of iteron containing copy-number-control systems. Such systems contain multiple repeats (iterons) that control plasmid copy-number at the origin of replication. Binding of a plasmid-encoded replication protein to the iterons is essential for initiation (42). In the P1, R6K and RK2 plasmids, iteron containing DNA molecules are paired by the cognate replication protein and the observed pairing was proposed to inhibit initiation of replication (43–45). In the parA partitioning system, binding of a plasmid-encoded protein to iterons mediates replicon pairing and this pairing may confer directionality to plasmid DNA segregation. Thus, mechanistically analogous DNA pairing reactions control different processes in prokaryotes.
Pairing (cohesion) of sister chromatids is a part of the mechanism of DNA segregation in eukaryotes. The role of chromatid cohesion is to constrain the kinetochores of the sister chromatids such that they attach to microtubules originating from both spindle poles. Upon release of the cohesion, the sister chromatids are pulled toward the opposite poles by the spindle apparatus (1–5). We propose that the function of plasmid pairing by the parA system is analogous. Symmetrical pairing of two DNA molecules can providing the dividing cell with the directionality required for ordered segregation of replicons. The paired plasmids could be recognized by an apparatus that moves the plasmid copies in opposite directions. Alternatively, the paired plasmids could attach to a cellular structure that positions the plasmid copies at opposite sides of the mid-cell septum.
The partitioning proteins Spo0J of B. subtilis, ParA and ParB of C. crescentus and SopB of the F plasmid have been found to localize toward the poles or at the cell quarter sites in predivisional cells (21, 23–25, 30). A bipolar localization is also observed for the ParM partitioning protein from plasmid R1 (R.B.J. and K.G., unpublished). For the B. subtilis chromosome and the F plasmid it is known that partitioning systems are involved in tethering the replicons to the specific positions in the cell (22, 28, 29). The function of specific localization of the partitioning proteins could be to tether replicons that already have been physically separated to positions in both daughter cells that are located far from the septum. In this model, the sister replicons are only paired in a part of the cell cycle before they are recognized by an apparatus that moves the replicons in opposite directions and tethers them to the bipolar positions. The similar genetic structure of the prokaryotic partitioning systems (i.e., two proteins and a centromere-like site that mediates incompatibility) suggests that replicon pairing before DNA segregation may occur in the process mediated by these systems as well.
Acknowledgments
This work was supported by the Danish Biotechnology Programme and the Plasmid Foundation. We thank Gerhild Lüder and Pia Hovendal for excellent technical assistance, Hansjörg Lehnherr for comments on the manuscript, Marie Gotfredsen for useful discussions and for donating purified ParR, and Dhruba Chattoraj for the suggestion to use the ligation assay.
References
- 1.Miyazaki W Y, Orr-Weaver T L. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- 2.Nicklas R B. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 3.Pluta A F, Mackay A M, Ainsztein A M, Goldberg I G, Earnshaw W C. Science. 1995;270:1591–1694. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 4.Hoyt M A, Geiser J R. Annu Rev Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- 5.Bickel S E, Orr-Weaver T L. BioEssays. 1996;18:293–300. doi: 10.1002/bies.950180407. [DOI] [PubMed] [Google Scholar]
- 6.Hiraga S. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 7.Wake R G, Errington J. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- 8.Abeles A L, Friedman S A, Austin S J. J Mol Biol. 1985;85:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- 9.Ogura T, Hiraga S. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes K, Molin S. J Mol Biol. 1986;190:269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- 11.Dam M, Gerdes K. J Mol Biol. 1994;236:1289–1298. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 12.Jensen R B, Dam M, Gerdes K. J Mol Biol. 1994;236:1299–1309. doi: 10.1016/0022-2836(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 13.Breüner A, Jensen R B, Dam M, Pedersen S, Gerdes K. Mol Microbiol. 1996;20:581–592. doi: 10.1046/j.1365-2958.1996.5351063.x. [DOI] [PubMed] [Google Scholar]
- 14.Jensen R B, Gerdes K. J Mol Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- 15.Davis M A, Austin S J. EMBO J. 1988;7:1881–1888. doi: 10.1002/j.1460-2075.1988.tb03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. J Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 17.Davis M A, Martin K A, Austin S J. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe E, Wachi M, Yamasaki M, Nagai K. Mol Gen Genet. 1992;234:346–352. doi: 10.1007/BF00538693. [DOI] [PubMed] [Google Scholar]
- 19.Ireton K, Gunther N W, IV, Grossman A D. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe M E, Errington J. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- 21.Mohl D A, Gober J W. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 22.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 23.Lin D C, Levin P A, Grossman A D. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 25.Lewis P J, Errington J. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- 26.Gordon G G, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 27.Hiraga S, Ichinose C, Niki H, Yamazoe M. Mol Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- 28.Lin D C, Grossman A D. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 29.Niki H, Hiraga S. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 30.Kim S-K, Wang J C. Proc Natl Acad Sci USA. 1998;95:1523–1527. doi: 10.1073/pnas.95.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler R T, Shapiro L. Cell. 1997;88:577–579. doi: 10.1016/s0092-8674(00)81898-x. [DOI] [PubMed] [Google Scholar]
- 32.Harry E J. Trends Microbiol. 1997;5:295–297. doi: 10.1016/S0966-842X(97)01091-3. [DOI] [PubMed] [Google Scholar]
- 33.Austin S, Abeles A. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- 34.Yanish-Perron C, Viera J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Martín J, del Solar G H, Lurz R, de la Campa A G, Dobrinski B, Espinosa M. J Biol Chem. 1989;264:21334–21339. [PubMed] [Google Scholar]
- 36.Mukhopadhyay G, Dibbens J A, Chattoraj D K. In: Methods in Molecular Genetics. Adolph K W, editor. Vol. 6. San Diego: Academic; 1995. pp. 400–420. [Google Scholar]
- 37.Lynch A S, Wang J C. J Mol Biol. 1994;236:679–684. doi: 10.1006/jmbi.1994.1179. [DOI] [PubMed] [Google Scholar]
- 38.Biek D P, Shi J. Proc Natl Acad Sci USA. 1994;91:8027–8031. doi: 10.1073/pnas.91.17.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biek D P, Strings J. J Mol Biol. 1995;246:388–400. doi: 10.1006/jmbi.1994.0094. [DOI] [PubMed] [Google Scholar]
- 40.Hanai R, Liu R, Benedetti P, Caron P R, Lynch A S, Wang J C. J Biol Chem. 1996;271:17469–17475. doi: 10.1074/jbc.271.29.17469. [DOI] [PubMed] [Google Scholar]
- 41.Austin S, Nordström K. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 42.Nordström K. Cell. 1990;63:1121–1124. doi: 10.1016/0092-8674(90)90405-4. [DOI] [PubMed] [Google Scholar]
- 43.Chattoraj D K, Mason R J, Wickner S H. Cell. 1988;52:551–557. doi: 10.1016/0092-8674(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 44.McEachern M J, Bott M A, Tooker P A, Helinski D R. Proc Natl Acad Sci USA. 1989;86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blasina A, Kittell B L, Toukdarian A E, Helinski D R. Proc Natl Acad Sci USA. 1996;93:3559–3564. doi: 10.1073/pnas.93.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]