Abstract
The stability of all single-base mismatched pairs between a peptide nucleic acid 11-mer and its complementary DNA has been quantified in terms of their melting temperature and compared with the limited amount of data published to date. The strength of the interaction was determined by an automated affinity-electrophoretic approach permitting the visualization, in real time, of hybridization between a physically immobilized peptide nucleic acid and a complementary DNA migrating in an electric field. The dissociation constants are in the range of 10−7 M (for mismatches) to 10−10 M (for fully complementary DNA), which are in excellent agreement with solution studies. These and other thermodynamic constants can be accurately, rapidly, and reproducibly measured in this system at concentrations approaching dissociation conditions by using fluorescently labeled DNA in conjunction with commercial DNA sequencers. The stability of single-base mismatched peptide nucleic acid–DNA duplexes depends both on the position as well as on the chemical nature of the mismatch. The stability is at a minimum when the mutation is positioned 4 bases from either terminus (a loss of 20°C or more in the melting temperature) but regains substantial stability when the mismatch is at the center of the duplex. The most stable mismatched pairs are G:T and T:T whereas destabilization is maximal for A:A and G:G. These observations are of significance in the design of probes for detecting mutations by hybridization.
Peptide nucleic acids (PNA) are synthetic chimeras of nucleobases linked to a peptide backbone (1). This spacing permits the bases to form, among other possible structures, standard base pairs with natural nucleic acids. The lack of the phosphodiester linkage, leading to an electronically neutral species, has, however, important consequences for the base-pairing potential of PNA. Investigations of the stability (2) and kinetics (3) of PNA–DNA (and –RNA) duplex formation have confirmed and quantified the existence of strong base-pairing interactions under various conditions and using different physical techniques ranging from calorimetry (2) to BIAcore measurements (3). However, there are considerable differences in the derived physical constants, which has led to the conclusion that the Kdiss values “determined so far for PNA–nucleic acid complexes must be considered most preliminary” (3) because the very high thermodynamic stability requires the use of concentrations that are at the lower detection limit of conventional techniques to achieve significant degrees of dissociation. The effect of isolated mismatch sites (3) is a destabilization of the duplex that can be applied usefully to the detection of mutations (4). The seemingly straightforward correlation between reduced stability and the loss of conventional Watson–Crick base pairing, which forms the basis of potential clinical applications, is marred by the occasional and unexplained remarkable stability of some “mismatches” (4). A systematic investigation relating the chemical and positional effect of mismatches on DNA–PNA duplex stability was, therefore, undertaken by extending the concept of affinity electrophoresis (5) to gain some insight into possible noncanonical base pair stabilization.
Affinity electrophoresis of macromolecules, depending on the noncovalent interaction of two species, requires the immobilization of one interacting partner in a gel matrix (5). Structure-specific affinity electrophoresis of RNA has been targeted at certain chemically defined regions of the molecule such as the termini or natural (6) and unnatural (7) modifications. Sequence-selective affinity electrophoresis of DNA has been attempted with some success by using certain intercalating reagents (8–10), but stringent base-pair-dependent sequence specificity is destined to failure in view of the irreconcilable requirement for high salt concentrations to achieve specific base pairing and the low salt requirement needed to permit electrophoresis.
The confirmed greater stability of PNA–nucleic acid duplexes, together with the salt independence of hybridization, has been ascribed to the lack of backbone charge repulsion. These two factors, namely electronic neutrality together with salt-independent base pair specificity, are the properties of PNA that make them ideal candidates as ligands in an affinity-electrophoretic partnership. The intrinsic sensitivity of affinity electrophoresis to environment-dependent structural changes (5) coupled with the extremely low detection limits of laser-induced fluorescence has been applied to PNA-containing gels. The concept of real-time hybridization is exemplified here by a quantitative study of the stability of base-paired duplexes.
MATERIALS AND METHODS
DNA primer R was synthesized by using an ABI394 synthesizer (Applied Biosystems) and bore a 5′-terminal fluorescent label by using FlourePrime phosphoramidite (Amersham–Pharmacia) during its synthesis. All other DNA oligonucleotides (Table 1) were synthesized by using an ABI3948 instrument (Applied Biosystems) in its auto-purification mode. They were labeled 3′-terminally with a single fluorescein-UTP with the aid of terminal transferase (Fermentas). Labeling was carried out by using 200 ng oligonucleotide/0.1 mM fluorescein ribo-UTP (NEN)/15 units terminal transferase in a total volume of 10 μl for 30 min at 37°C, as described (11). The reaction was stopped by heating to 95°C for 2 min, and the labeled product was diluted as desired directly from the reaction mixture. PNA was custom-made by PerSeptive Biosystems (Framingham, MA). It was purified by gel filtration over a NAP5 column (Amersham–Pharmacia) in water, collecting 0.5-ml fractions. Fractions containing the UV-absorbing peak were pooled and stored frozen. Affinity gels were constructed by mixing 9 ml 2× MDE (FMC), 3.6 ml 10× TBE (10× TBE: 1 M Tris/0.83 M boric acid/10 mM EDTA) together with the appropriate amount of PNA, as indicated in a total volume of 36 ml. Polymerization was initiated by the addition of 130 μl 10% ammonium persulfate and 30 μl Temed. The gel solution was poured immediately between the plates of an ALF DNA sequencer (Amersham–Pharmacia) with the use of 0.35-mm spacers. The temperature of the gel was controlled externally by connecting a MultiTempII thermostat bath (Amersham–Pharmacia) to the ALF thermostat plate. Gels were run routinely in 0.8× TBE at 34-W constant power. Fluorescent samples (≈7.5 nM) were applied in 1% Dextran blue/formamide after denaturing at 95°C for 3 min and quenching in ice. The migration pattern was quantified by using fragmentmanager 1.2 (Amersham–Pharmacia) and analyzed graphically in Microsoft excel 8.0.
Table 1.
Oligonucleotide and PNA sequences
| Name | Sequence | mm posn. | Tm, °C |
|---|---|---|---|
| Single mismatched oligonucleotides | |||
| mmgt | 5′ ggaaaTaccaatgatat-Fu 3′ | 1 | 45.1 |
| G | |||
| mm1 | 5′ ggaaaGaccaatgatat-Fu 3′ | 1 | 42.5 |
| G | |||
| mm19 | 5′ ggaaaAaccaatgatat-Fu 3′ | 1 | 42.5 |
| G | |||
| mm20 | 5′ ggaaacGccaatgatat-Fu 3′ | 2 | 39.3 |
| T | |||
| mm35 | 5′ ggaaacCccaatgatat-Fu 3′ | 2 | 47.4 |
| T | |||
| mm2 | 5′ ggaaacTccaatgatat-Fu 3′ | 2 | 39.0 |
| T | |||
| mm3 | 5′ ggaaacaGcaatgatat-Fu3′ | 3 | 27.5 |
| G | |||
| mm21 | 5′ ggaaacaAcaatgatat-Fu 3′ | 3 | 29.4 |
| G | |||
| mm22 | 5′ ggaaacaTcaatgatat-Fu 3′ | 3 | 36.2 |
| G | |||
| mm23 | 5′ ggaaacacAaatgatat-Fu 3′ | 4 | 23.2 |
| G | |||
| mm24 | 5′ ggaaacacTaatgatat-Fu 3′ | 4 | 35.0 |
| G | |||
| mm4 | 5′ ggaaacacGaatgatat-Fu 3′ | 4 | 24.6 |
| G | |||
| mm5 | 5′ ggaaacaccTatgatat-Fu 3′ | 5 | 41.0 |
| T | |||
| mm14 | 5′ ggaaacaccCatgatat-Fu 3′ | 5 | 37.1 |
| T | |||
| mm25 | 5′ ggaaacaccGatgatat-Fu 3′ | 5 | 37.1 |
| T | |||
| mm6 | 5′ ggaaacaccaTtgatat-Fu 3′ | 6 | 39.6 |
| T | |||
| mm13 | 5′ ggaaacaccaCtgatat-Fu 3′ | 6 | 32.8 |
| T | |||
| mm26 | 5′ ggaaacaccaGtgatat-Fu 3′ | 6 | 35.4 |
| T | |||
| mm7 | 5′ ggaaacaccaaAgatat-Fu 3′ | 7 | 31.3 |
| A | |||
| mm12 | 5′ ggaaacaccaaGgatat-Fu 3′ | 7 | 32.3 |
| A | |||
| mm27 | 5′ ggaaacaccaaCgatat-Fu 3′ | 7 | 29.5 |
| A | |||
| mm28 | 5′ ggaaacaccaatAatat-Fu 3′ | 8 | 30.0 |
| C | |||
| mm29 | 5′ ggaaacaccaatTatat-Fu 3′ | 8 | 31.2 |
| C | |||
| mm8 | 5′ ggaaacaccaatCatat-Fu 3′ | 8 | 27.9 |
| C | |||
| mm9 | 5′ ggaaacaccaatgTtat-Fu 3′ | 9 | 43.7 |
| T | |||
| mm30 | 5′ ggaaacaccaatgGtat-Fu 3′ | 9 | 41.0 |
| T | |||
| mm34 | 5′ ggaaacaccaatgCtat-Fu 3′ | 9 | 40.7 |
| T | |||
| mm10 | 5′ ggaaacaccaatgaAat-Fu 3′ | 10 | 42.5 |
| A | |||
| mm31 | 5′ ggaaacaccaatgaGat-Fu 3′ | 10 | 43.5 |
| A | |||
| mm32 | 5′ ggaaacaccaatgaCat-Fu 3′ | 10 | 43.5 |
| A | |||
| mm11 | 5′ ggaaacaccaatgatTt-Fu 3′ | 11 | 48.1 |
| T | |||
| mm33 | 5′ ggaaacaccaatgatGt-Fu 3′ | 11 | 48.5 |
| T | |||
| mm36 | 5′ ggaaacaccaatgatCt-Fu 3′ | 11 | 38.5 |
| T | |||
| 430 | 5′ ggaaacaccaatgatat-Fu 3′ | None | 48.9 |
| Multiple mismatched oligonucleotides | |||
| R | 5′ F-caggaaacagctatgacc | NA | NA |
| 429 | 5′ aacaccaaAgatGatat-Fu 3′ | 7,11 | 29.0 |
| A T | |||
| 431 | 5′ atatcatctttggtgtt-Fu 3′ | mult | ** |
| mm15 | 5′ ggaaacaccTatgTtat-Fu 3′ | 5,9 | 28.1 |
| T T | |||
| mm16 | 5′ ggaaacCccTatgCtat-Fu 3′ | 2,5,9 | 21.7 |
| T T T | |||
| mm17 | 5′ ggaaacaccTatgatCt-Fu 3′ | 5,11 | 36.6 |
| T T | |||
| mm18 | 5′ ggaaacTccTTtgTtTt-Fu 3′ | mult | ** |
| T TT T T | |||
| PNA | C- gtggttactat-O N | ||
The PNA sequence is shown in its antiparallel form to facilitate a visualization of the complementary oligonucleotide sequences (shown in bold). The 3′-terminal fluorescent UTP is denoted by Fu and the PNA N-terminal linker is shown by O. Mismatched bases are given in uppercase letters with the corresponding base on the PNA strand shown below them. The melting temperatures, determined as described in Materials and Methods, at 100 nM immobilized PNA are indicated. The reproducibility was ±0.5°C. Tm values below 25°C were estimated by graphic extrapolation of the melting curves. No detectable interaction is denoted by ∗∗. The position of the mismatches, starting at the 5′ end of the DNA strand, are given (mm posn.). More than two mismatched bases are designated mult. NA, not applicable.
RESULTS
Entrapment of PNA in an Electrophoresis Gel Causes Retardation of a Complementary Oligonucleotide.
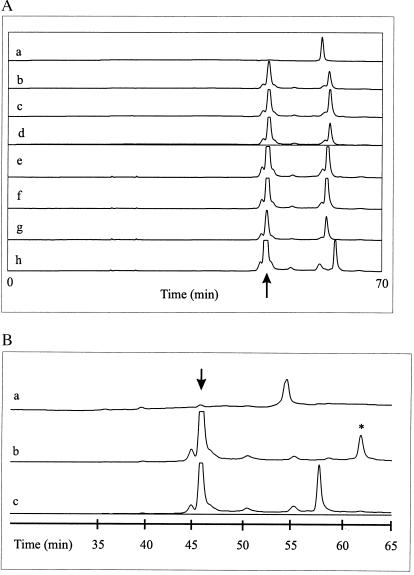
In an unmodified, native 0.5× MDE gel, all fluorescein-labeled oligonucleotides (18 bases plus 3′ fluorescein) are virtually indistinguishable but migrate slightly slower than the reference universal reverse primer R (17 bases plus 5′ fluorescein), as exemplified in Fig. 1A, but slightly faster than oligonucleotide 431, reflecting a completely different base composition of the latter (Table 1). The relative migration difference with respect to oligonucleotide R is independent of temperature (not shown) and is, therefore, characteristic for each oligonucleotide under conditions where there is zero sequence-specific interaction with the matrix. Addition of a PNA 11-mer to the gel at a concentration of 100 nM before polymerization leads to entrapment of the PNA and to a marked retardation of the fully complementary 430 with respect to R (Fig. 1B). The retardation represents a shift of 10% or more in the migration distance, corresponding to a separation of 5 min or more from an unretarded component. Oligonucleotide 431 being noncomplementary to PNA is unaffected by the presence of PNA and has a migration behavior with respect to oligonucleotide R as in a gel lacking PNA (Fig. 1A).
Figure 1.
Migration of fluorescent oligonucleotides as monitored by the detection system of an ALF DNA sequencer at 40°C. (A) Unmodified 0.5× MDE gel (no PNA). Approximately 7.5 nM (3 μl) of oligonucleotide R (a), mm1 (b), mm2 (c), mm4 (d), mmgt (e), 429 (f), 430 (g), and 431 (h) were applied, as described in Materials and Methods. (B) PNA (100 nM) was included in the gel. The applied samples were R (a), fully complementary 430 (b), and unrelated 431 (c). Arrows indicate the position of residual excess fluorescein-UTP from the labeling reaction. The retarded signal in B is emphasized by an asterisk.
Determination of the Melting Temperature (Tm) of PNA–DNA Duplexes as a Function of PNA Concentration.
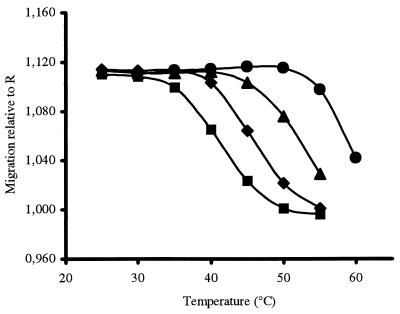
The relative retardation of the interacting oligonucleotides depends on the temperature of the gel at any given concentration of entrapped PNA; the interaction becomes weaker at higher temperatures and the retardation is correspondingly reduced. Thus, a plot of retardation (with respect to the migration of primer R) as a function of temperature is of a sigmoid nature, resembling a typical melting profile of a nucleic acid duplex (Fig. 2).
Figure 2.
Temperature dependence of the interaction of the fully complementary oligonucleotide 430 with the matrix-bound PNA as a function of its concentration. The relative migration, in terms of data points collected to the midpoint of the detected peak, was determined with respect to the noninteracting oligonucleotide R. PNA was included at a concentration of 1,000 nM (•), 200 nM (▾), 20 nM (♦), and 5 nM (■).
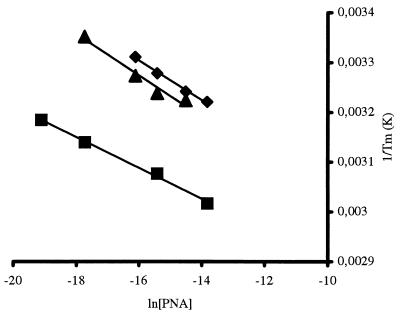
The Tm can be deduced from such curves as the point of 50% maximal retardation. Furthermore, as would be expected from the theory of duplex formation (12), the Tm depends on the concentration of PNA. Thus, a series of electrophoretic profiles at varying PNA concentrations gave rise to a set of Tm values whose dependence on the concentration of the immobilized partner could be used to calculate the corresponding thermodynamic constants from a plot of 1/Tm vs. ln[PNA] (13) (Fig. 3). The linear relationship provides the slope = R/ΔH° and an intercept = (ΔS° − Rln4)/ΔH°, from which ΔG° and Kdiss may be derived (Table 2).
Figure 3.
Relationship between the Tm determined from curves such as those depicted in Fig. 2 and the concentration of the interacting PNA. The analysis is exemplified for the fully complementary oligonucleotide 430 (■), single mismatch mm13 (▾), and double mismatch 429 (♦).
Table 2.
Thermodynamic parameters for DNA–PNA duplex formation at 37°C
| DNA | Mismatch position and type | ΔS°, eu | ΔH°, kcal/mol | ΔG°(37°C), kcal/mol | Kdiss (37°C), M |
|---|---|---|---|---|---|
| 430 | None | −164 | −64 | −14.7 | 3.1 × 10−10 |
| mm1 | 1 (G:G) | −122 | −50 | −11.9 | 4.2 × 10−9 |
| mmgt | 1 (G:T) | −135 | −54 | −12.1 | 3.2 × 10−9 |
| mm2 | 2 (T:T) | −129 | −51 | −11.2 | 1.3 × 10−8 |
| mm3 | 3 (G:G) | −179 | −64 | −8.8 | 6.3 × 10−7 |
| mm4 | 4 (G:G) | −213 | −74 | −8.0 | 2.3 × 10−6 |
| mm5 | 5 (T:T) | −142 | −55 | −11.5 | 8.4 × 10−9 |
| mm6 | 6 (T:T) | −138 | −54 | −11.2 | 1.2 × 10−8 |
| mm13 | 6 (C:T) | −121 | −47 | −10.8 | 1.0 × 10−7 |
| mm7 | 7 (A:A) | −261 | −90 | −9.9 | 1.0 × 10−7 |
| mm12 | 7 (G:A) | −169 | −62 | −9.9 | 1.0 × 10−7 |
| mm8 | 8 (C:C) | −88 | −37 | −9.6 | 1.6 × 10−7 |
| mm9 | 9 (T:T) | −123 | −50 | −11.8 | 4.8 × 10−9 |
| mm10 | 10 (A:A) | −103 | −43 | −11.3 | 1.1 × 10−8 |
| mm11 | 11 (T:T) | −138 | −55 | −12.6 | 1.3 × 10−9 |
| 429 | 7 (A:A), 11 (G:T) | −130 | −50 | −9.5 | 2.3 × 10−7 |
eu, cal K−1·mol−1.
Mismatches Destabilize the Duplex.
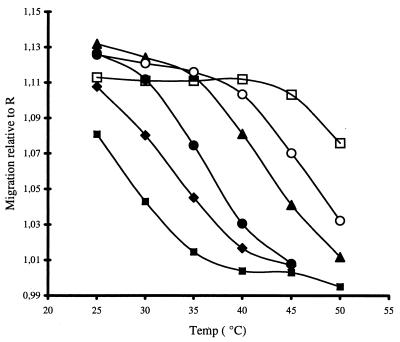
Single-base as well as multiple mismatches reduce the stability of the duplexes. The temperature dependence of the retardation is shifted accordingly to lower Tm values (Fig. 4). The PNA concentration dependence of the Tm was determined for a set of oligonucleotides having a single-base mismatch at each position, as described above, and the thermodynamic parameters were calculated, as summarized in Table 2.
Figure 4.
Temperature dependence of the interaction of the fully complementary oligonucleotide 430 (□) with the matrix-bound PNA (200 nM) compared with single mismatch probes: mm9 (○), mm2 (▾), mm12 (•), mm8 (♦), and mm4 (■). The relative migration, in terms of data points collected to the midpoint of the detected peak, was determined with respect to the noninteracting oligonucleotide R.
The Duplex Stability Depends on the Position and the Nature of the Single Mismatch.
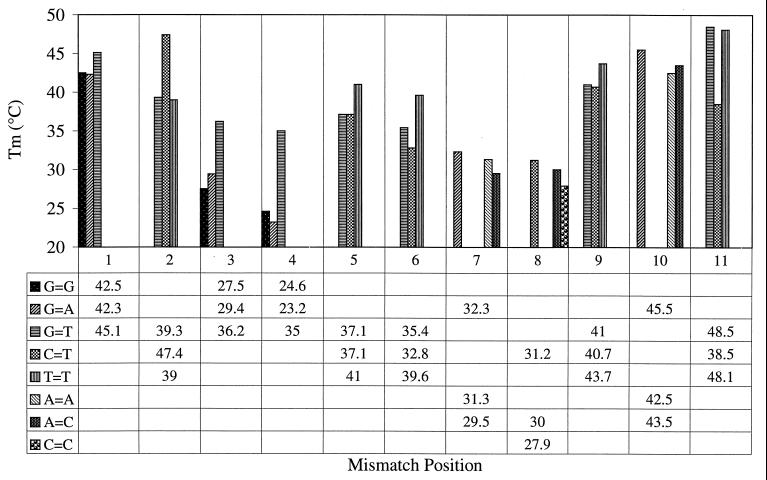
By systematically screening all possible single-base mismatches at each position in the 11-mer region of the DNA corresponding to the PNA (summarized in Table 1) at a single PNA concentration (100 nM), one could rapidly establish a correlation between the stability (in terms of the Tm) and the position of the mismatch. The results of this survey are presented graphically in Fig. 5. An overall loss of stability is seen on moving the mismatch away from the termini, but interestingly, it is also observed that certain noncanonical base interactions have a stabilizing influence at positions that otherwise weaken duplex formation. Multiple mismatched base pairs are, in general, significantly less stable than single mismatches (Table 1, Multiple mismatched oligonucleotides), although some quite stable combinations (e.g., mm17) were noted.
Figure 5.
Stability of single-mismatch base pairs as a function of their position in the PNA. The Tm for the mismatches indicated at each position is depicted. The melting temperatures shown in the table were obtained at 100 nM PNA.
DISCUSSION
The interaction between a DNA strand and its complementary PNA has been studied by a variety of techniques (2–4) and been quantified as leading to the formation of thermally more stable duplexes than is the case with natural nucleic acids. A consequence of this stability is, among other effects, an enhanced discrimination between fully complementary and mismatched duplexes because of a greater reduction in Tm for single mismatched DNA–PNA pairs compared with the corresponding DNA–DNA duplex (1). This has led to the development of such applications of PNA–DNA hybridization where the specificity of detection is of primary consideration (4). For such purposes, any unforeseen sites or structural hot spots encouraging stable mismatches would have serious consequences. Indeed, reports describing unexpectedly stable hybridization signals (4) create a question mark over a potentially significant advance in the collection of sequence-specific information.
In an attempt to examine systematically the effects of the position and nature of mismatches on the stability of DNA–PNA duplexes, all possible single-base mismatches of a DNA 11-mer were synthesized and their interaction with the corresponding PNA was quantified. In contrast to the physical methods used previously for measuring duplex stability [calorimetry, hyperchromicity (2), BIAcore (3)], quantitative real-time hybridization in the form of affinity electrophoresis has been introduced. An affinity matrix composed of a polyacrylamide gel containing a physically entrapped PNA has been shown to retard significantly and specifically only such DNA molecules that are related to the PNA through complementarity. The extremely high binding affinity between DNA and PNA that has hampered the measurement of thermodynamic parameters by, for example, the BIAcore technique (3) allows the use of low concentrations (in the nM range) of PNA in the gel, which, on the one hand, permits an economical use of the PNA and, more significantly, causes no distortion or other disruption of the electrophoresis system. Sensitivity is maintained by monitoring the migration of a fluorescently labeled DNA sample through the gel. Data may be collected conveniently by any commercial, gel-based, automated DNA sequencer. These affinity gels may, for qualitative purposes, be reused several times even after storage within the sequencer for several days. The retardation of an interacting DNA strand with the immobilized PNA is highly reproducible (data not shown) and sufficiently separated from an internal, noninteracting control DNA to permit reliable analysis. Furthermore, as would be expected for double-strand formation, the retardation is temperature-dependent, reflecting a thermal equilibration process. In a rigorous quantification of binding processes one needs to differentiate between the amount of PNA that has been included in the polymerization mixture and the quantity of PNA that is subsequently available for interaction. Physical losses are limited to the minimal amounts that diffuse from the edges of the gel during polymerization and electrophoresis. PNA, being electronically neutral, does not migrate during electrophoresis (13). Chemical degradation of PNA during the radical-induced polymerization is unlikely. The major bases of nucleic acids do not appear to be targets for radical attack (6) and, although phosphodiester bonds are susceptible to radical-induced cleavage (14), it is this particular bond that has been replaced in PNA by the amide linkage. On the other hand, amide bonds are already present in the form of acrylamide in an excess of several orders of magnitude over the PNA, so that the latter would be well screened from radical attack. One may speculate whether all the physically entrapped PNA in the gel is available for interaction with the mobile DNA. A definitive answer would require an independent determination of the identical thermodynamic parameters. However, the given interpretation of the observed phenomenon, equating concentration with molecular activity, is supported by the fact that when the duplex melting temperature obtained by this strategy is analyzed in terms of established thermodynamic principles (15), a linear dependence on the entrapped PNA concentration is observed. Furthermore, comparison of the data with the literature values from solution studies is in excellent agreement with comparable interactions. Thus, Tomac et al. (2) reported figures for a 10-mer PNA of ΔH° −63 kcal/mol, ΔS° −168 eu, ΔG° −13 kcal/mol (298 K) (cf. results from Table 2, above). One would appear, therefore, to be justified in considering the affinity-electrophoretic procedure to reflect an interaction in solution rather than being of the type seen at solid surfaces (3) that has given values of Kdiss 6–7 orders of magnitude higher, i.e., weaker binding, than measured here.
Other factors that destabilize the overall DNA–PNA structure are readily detected by this method. Of significance for this investigation has been the sensitivity of the interaction to single-base mismatches. Thermodynamic parameters were determined for mismatches at every position of the duplex and compared with the wild-type sequence. Although they provided thermodynamic data relevant to the destabilization, these results were not able to provide information as to the positional effect of the mismatch because a secondary chemical effect could not be ruled out, i.e., both the position and the bases involved in the mismatch were altered. To obtain a more systematic view all 33 possible mismatches were screened by determining their Tm at 100 nM PNA. A graphical representation of these results (Fig. 5) leads to the following conclusions. As would be expected intuitively, mismatches at or adjacent to the termini have little effect but the stability of the duplex decreases as the mismatch moves away from the terminus (irrespective of whether 5′ or 3′). Mutations near the center of the duplex regain a certain stability, which may be because of the formation of short (4–5 bases) duplex stretches at either end. The sum of this stabilization then would have to be greater than that of a fully formed 7-bp duplex with a weakly held terminal triplet (mismatch at position 4). The extent of this effect could be expected to be dependent on the base pairs involved in the longer “tail” duplex. With the present sequence, loss of a total of 3 G:C and 1 A:T base pairs, as in mm4 at the 5′ end, apparently has more severe consequences on the stability than dissolution of 3 A:T and 1 G:C interactions at the 3′ end in mm8. Whether this effect is specific to this system or can be extended to a general weakening of centrally mismatched duplexes remains to be seen. The relatively short segment of DNA that has been studied systematically here does not permit an absolute statement about the influence of context on mismatch stability. DNA–DNA mismatched base pairs have been the subject of intensive study, mostly by NMR. The information gained from these investigations cannot be compared rigorously with the results presented here. First, it is in the nature of NMR that substantially higher concentrations of the components are involved, and second, and more important, the flexible, uncharged nature of the PNA backbone would be expected to permit a wider range of conformations than is possible for the phosphodiester chain (1). Thus, although not disputing the general finding that the stability of mismatches in DNA–DNA helixes is governed largely by sequence context (16–20), other stacking and conformational factors may be available for DNA–PNA duplexes. Additionally, the role of water molecules as noted for G:T base pairs in DNA–DNA (21) and possible H bonding with the amide backbone of PNA complicates the analysis. It is nevertheless notable that T:T mismatches are found to offer the same or greater stability than potentially wobble-base-paired G:T interactions (Fig. 5). In DNA–DNA complexes, the imino-carbonyl H bond of T:T was found, by NMR, to be weaker than in G:T (22), although Gervais et al. (23) detected the formation of two such interactions in the T:T structure. The stability of mismatches at the center of DNA–PNA duplexes has been reported previously from BIAcore studies (3). The single T:T mismatch described by these authors also had the same effect on the Tm value as a T:G base pair. Perhaps of particular significance in this respect are the mismatched sequences giving extremely stable hybridization signals in a PNA array investigation (4). The mismatches in question were single T:T, double AT:AT, and triple TAT:TAT mismatched sequences. However, it would seem likely that other factors, apart from the presence of T:T interactions, are involved in the documented stability. Merely increasing the number of T:T mismatches (or mismatches, in general) leads to steady reduction in stability (Table 1, Multiple mismatched oligonucleotides). Single T:T mutants mm5 (Tm = 41°C) and mm9 (Tm = 43.7°C) give rise to double T:T at the same positions, mm15, with Tm = 28.1°C and all T:T mm18, which showed no binding at all. The effect of sequence context then must also play a role in these interactions and could also, in part, explain the tremendous loss in stability of up to 25°C in Tm of certain single-base mismatches such as at position 4. A practical consequence of these measurements is that one cannot, in general, assume that mismatches at the center of a probe are more discriminating than those nearer the termini. The design of hybridization probes should, therefore, take into account not only the nature of the mismatched base pair itself, but also the stability and context of the residual sequences at either side of the mutation.
The sequence-specific hybridization of nucleic acids has been of fundamental importance in supporting many major advances in molecular biology over the past two decades. Although a range of modifications and technical developments have increased the scope and fields of application, the basic principle has remained unaltered. With the increasing demand on the rapid availability of genetic analysis, conventional hybridization techniques soon reach a practical limit. Advances in chip technology promise to alleviate problems associated with capacity but, as has been noted (4), are not yet entirely free of other drawbacks in the form of sensitivity and selectivity. Furthermore, use of arrays does not necessarily address the problem of speed because a period of hybridization, washing, and detection must precede data analysis. Real-time hybridization during electrophoretic fractionation has not been at the forefront of possible alternatives for a variety of technical reasons. The main hindrance has been the strong salt dependence of specific base pairing, which is irreconcilable with electrophoretic techniques. However, advances in the construction of uncharged DNA analogs, in particular, PNA, which form highly stable, specific duplexes with DNA (and RNA) at low salt concentrations, could form the basis of novel electrophoretic applications.
It now has been demonstrated that PNA can be entrapped physically in a gel matrix in such a way as to permit specific hybridization during the electrophoretic passage of a complementary DNA. A constellation of this type—an immobilized ligand interacting with a mobile partner under the influence of an electric field—has been defined previously as an affinity-electrophoretic system (5) and offers all the advantages of such analyses. Additionally, the use of fluorescence detection, for instance, as available in commercial DNA sequencers, provides an opportunity for analyzing in real time a reasonable number of samples automatically and at extreme sensitivity. Other applications, both preparative and high-capacity analytical techniques, based on a multiplex-type of use of several DNA species labeled with different dyes but analyzed simultaneously in gels containing the corresponding mixed PNA partners, are envisaged.
Acknowledgments
I am grateful to Mrs. Elfi Scheifermayr for dedicated technical assistance. Financial support was given, in part, by the Deutsche Forschungsgemeinschaft (SFB388).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PNA, peptide nucleic acid; Tm, melting temperature.
References
- 1.Eriksson M, Nielsen P E. Q Rev Biophys. 1996;29:369–394. doi: 10.1017/s0033583500005886. [DOI] [PubMed] [Google Scholar]
- 2.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen P E, Nordén B, Gräslund A. J Am Chem Soc. 1996;118:5544–5552. [Google Scholar]
- 3.Jensen K K, Ørum H, Nielsen P E, Nordén B. Biochemistry. 1997;36:5072–5077. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]
- 4.Weiler J, Gausepohl H, Hauser N, Jensen O N, Hoheisel J D. Nucleic Acids Res. 1997;25:2792–2799. doi: 10.1093/nar/25.14.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Igloi G L. In: Molecular Interactions in Bioseparations. Ngo T T, editor. New York: Plenum; 1993. pp. 511–531. [Google Scholar]
- 6.Igloi G L. Biochemistry. 1988;27:3842–3849. doi: 10.1021/bi00410a048. [DOI] [PubMed] [Google Scholar]
- 7.Igloi G L. Anal Biochem. 1992;206:363–368. doi: 10.1016/0003-2697(92)90379-l. [DOI] [PubMed] [Google Scholar]
- 8.Takai H, Yamakowa H, Ohara O, Sakaguchi-Inoue J. BioTechniques. 1997;23:58–60. doi: 10.2144/97231bm10. [DOI] [PubMed] [Google Scholar]
- 9.Müller M, Kruse L, Tabrett A M, Barbara D J. Nucleic Acids Res. 1997;25:5125–5126. doi: 10.1093/nar/25.24.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yashima E, Suehiro N, Miyauchi N, Akashi M. J Chromatogr A. 1993;654:151–158. doi: 10.1016/0021-9673(93)83075-4. [DOI] [PubMed] [Google Scholar]
- 11.Igloi G L. Methods Mol Biol. 1996;65:23–28. doi: 10.1385/0-89603-344-9:23. [DOI] [PubMed] [Google Scholar]
- 12.Cantor C R, Schimmel P R. Biophysical Chemistry. San Francisco: Freeman; 1980. [Google Scholar]
- 13.Perry-O’Keefe H, Yao X-W, Coull J M, Fuchs M, Egholm M. Proc Natl Acad Sci USA. 1996;93:14670–14675. doi: 10.1073/pnas.93.25.14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perrin D M, Mazumder A, Sigman D S. Prog Nucleic Acids Res Mol Biol. 1996;52:122–151. doi: 10.1016/s0079-6603(08)60966-6. [DOI] [PubMed] [Google Scholar]
- 15.Marky L A, Breslauer K J. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 16.Allawi H T, SantaLucia J., Jr Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J W, Chou S H, Reid B R. J Mol Biol. 1992;228:1037–1041. doi: 10.1016/0022-2836(92)90312-8. [DOI] [PubMed] [Google Scholar]
- 18.Greene K L, Jones R L, Li Y, Robinson H, Wang A H, Zon G, Wilson W D. Biochemistry. 1994;33:1053–1062. doi: 10.1021/bi00171a003. [DOI] [PubMed] [Google Scholar]
- 19.Ke S H, Wartell R M. Nucleic Acids Res. 1993;21:5137–5143. doi: 10.1093/nar/21.22.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharyya A, Lilley D M. J Mol Biol. 1989;209:583–597. doi: 10.1016/0022-2836(89)90596-2. [DOI] [PubMed] [Google Scholar]
- 21.Kneale G, Brown T, Kennard O, Rabinovich D. J Mol Biol. 1985;186:805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- 22.Kouchakdjian M, Li B F, Swann P F, Patel D J. J Mol Biol. 1988;202:139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 23.Gervais V, Cognet J A, LeBret M, Sowers L C, Fazakerly G V. Eur J Biochem. 1995;228:279–290. [PubMed] [Google Scholar]