Abstract
Background
A great deal of evidence has shown that non‐human leucocyte antigen (HLA)‐B27 genes may play crucial roles in the aetiology of ankylosing spondylitis (AS), but there is little evidence of a relationship with tumour necrosis factor (TNF)α gene variation. One functional single‐nucleotide polymorphism (SNP), −850 C→T, on the TNFα gene promoter region was identified and confirmed to be significantly associated with AS by our recent study.
Objective
To investigate whether the −850 C→T SNP is a susceptibility locus for AS or is only a marker linked to potential disease gene loci in a Chinese population.
Methods
Ten common SNPs were selected from nine inflammatory genes covering the right and left flanking regions of the TNFα gene, which span a region of about 100 kb on chromosome 6p21.31, and a tag SNP in HCP5 gene was used to examine the linkage between the HLA‐B27 and TNFα genes. SNPs were genotyped by PCR restriction‐fragment length polymorphism (RFLP), allele‐specific PCR and restriction site‐generating PCR‐RFLP for single‐base association and linkage disequilibrium (LD).
Results
The prevalence of TNFα‐850 C→T SNP was significantly different between case and control groups. A specific haplotype covering TNFα gene mutant was strongly associated with AS. An LD test showed that a recombination between HLA‐B27 and TNFα might have taken place.
Conclusion
The TNFα locus was reconfirmed and showed association with susceptibility to AS. It may be independent of HLA‐B27. A range of 58 kb covering TNFα had strong LD to AS.
Keywords: TNFα , ‐850, association, ankylosing spondylitis
TNFα is a multifunctional pre‐inflammatory factor that has been implicated in many autoimmune diseases including type 1 diabetes, rheumatoid arthritis and ankylosing spondylitis (AS). The TNFα gene has been mapped to chromosome 6p21.31, with which a strong linkage to AS has been reported by several genome‐wide screening studies in diverse populations. Recently, studies have indicated that anti‐TNFα therapy is highly effective in AS patients and obtained significant and sustained improvement in inflammatory reactions. 1,2,3 However, whether the TNFα gene increases the risk of developing AS is still unclear.
To clarify whether TNFα gene is involved in the genetic aetiology of AS, we sequenced the entire promoter sequence of TNFα gene in samples from 36 patients with AS from the Shantou region in southern China. We found and reported a high frequency of variation of the –850C→T allele (rs1799724), which was strongly associated with AS in a case–control study (p<0.001).4 We then performed a similar study in a population from Yinchuan in the northwest of China, with similar results (p<0.001). We therefore considered that the TNFα gene variation might be a novel allele associated with AS, which has not been reported previously. Thus, to reconfirm this possible association, we replicated the study in a third remote population from the northeast of China, in Jilin province.
In addition, even if we were to confirm that the –850T allele is strongly associated with AS, the question of whether the variation is a cause of disease susceptibility or is only a marker closely linking to true disease loci remained to be answered. Thus, to attempt to map genes for AS susceptibility to a limited genomic area, we intensively screened a region of 100 kb covering the right and left flanking areas of the TNFα gene on chromosome 6p21.31 for single allele‐based haplotype association analysis and haplotype‐based LD test in another population.
Methods
The research ethics committee of Beijing Hospital approved this study. Informed consent was obtained from all subjects involved, and the study was performed with compliance with all principles of the Helsinki Accord.
Participants
In total, 79 unrelated patients with AS were enrolled in the study. All were attending the outpatient rheumatology department of the Affiliated Hospital of Beihua University School of Medicine. The diagnosis of AS strictly conformed to the modified New York criteria.5 In addition, 132 healthy unrelated blood donors were recruited from Jilin province.
Single‐nucleotide polymorphism selection
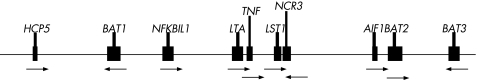
We selected nine SNPs, exclusive of rs1799724, from eight inflammatory genes located in a 100 kb region spanning the right and left sides of the TNFα gene. Of these nine SNPs, Of these rs2071592, rs909253 and rs1041981 have been reported to have strong association with inflammation and autoimmune disorders.6,7 The SNPs rs2284178, rs2256965, rs2242660 and rs2736191 were tag SNPs identified by the HapMap database (www.hapmap.org), and the SNPs rs929138 and rs3117583 were randomly selected. The SNP rs2284178 was used to detect the linkage between the TNFα and the human leucocyte antigen B27 (HLA‐B27) genes. The chromosome distribution of the genotyped SNPs are shown in fig 1.
Figure 1 Target gene alignments in the human leucocyte antigen class III region on chromosome 6.
PCR and allele‐specific PCR
HLA‐B27 typing of AS patients was performed by flow cytometry and that of healthy controls were fulfilled by PCR as described previously.8 The SNP rs1041981 was typed by allele‐specific PCR.
Restriction site‐generating PCR fragment length polymorphism analysis
The TNF850C→T SNP (rs1799724) was typed by semi‐nested PCR and restriction site‐generating PCR (RG‐PCR). The forward primer F1 and the reverse primer amplified a larger PCR product used as the template for semi‐nested PCR, then the forward primer F2, which carried a C→G single base substitution on the 3′ terminal to produce a Hpa I restriction site, was used with the reverse primer in a second PCR reaction.
The rs224660, rs2071592and rs929138 SNPs were genotyped by RG‐PC using a T→A base change on the 3′ terminal of the rs224660 reverse primer to form a AluI restriction site, and by producing base changes of C→A and G→C on the 3′ terminal of the rs2071592 and rs929138 forward primers to introduce a TaqI and PvuII restriction site, respectively.
PCR‐RFLP
The SNPs rs2284178, rs909253, rs2256965, rs2736191, rs2857697 and rs3117583 were typed by PCR‐RFLP. The restriction endonucleases used were HaeIII, NcoI, AvaII, PstI, MspI, and ApaI, respectively (the sequences of primers for typing 11 SNPs can be found in supplementary table 1; available at http://jmg.bmj.com/supplemental).
Table 1 Demographic and clinical characteristics of patients with AS (n = 79).
| Characteristic | Frequency |
|---|---|
| Age (years), mean (SD) (range)* | 32.2 (10.4) (14 to 58) |
| Women (%) | 35.4 |
| HLA‐B27 positive (%) | 89.9 |
| Peripheral arthritis (%) | 28.9 |
*Because clinical data on the age at first presentation and age at diagnosis were incomplete, they were not included in table 1.
Statistical analysis
Genotype frequencies were tested for Hardy−Weinberg equilibrium (HWE) by χ2 test (see supplementary file 1; available at http://jmg.bmj.com/supplemental). A two‐sided p value of <0.05 was considered significant. Single allele‐based association tests were performed using SPSS V.10.0 software (SPSS Inc., Chicago, IL, USA). Haplotype analysis, LD testing and haplotype‐based association studies were performed using Haploview V.3.32 software.9
Results
The characteristics of the 79 patients with AS are summarised in table 1. The distribution of genotypes between cases and controls for all SNPs are shown in table 2, and the results of haplotype analysis are shown in table 3.
Table 2 Analysis of SNPs: genotype and alleles distribution between AS and control groups.
| Genotype | AS, n = 79 | Controls, n = 132 |
|---|---|---|
| HCP5 (rs2284178) | ||
| CC | 22 (27.9%) | 34 (25.8%) |
| CT | 41 (51.9%) | 88 (66.7%) |
| TT | 16 (20.2%) | 10 (7.5%) |
| χ2 (p value) CT+TT vs CC | 0.11 (0.74) | |
| OR (95% CI) | 0.90 (0.48 to 1.68) | |
| BAT1 (rs929138) | ||
| AA | 32 (40.5%) | 64 (48.5%) |
| AG | 41 (51.9%) | 62 (47%) |
| GG | 6 (7.6%) | 6 (4.5%) |
| χ2 (p value) AG+GG vs AA | 1.27 (0.26) | |
| OR (95% CI) | 1.38 (0.78 to 2.43) | |
| NFKBIL‐1 (rs2071592) | ||
| AA | 28 (35.4%) | 49 (38.3%) |
| AT | 4050.6%) | 57 (44.5%) |
| TT | 11 (14.0%) | 22 (17.2%) |
| χ2 (p value) TT+AT vs AA | 0.17 (0.89) | |
| OR (95% CI) | 0.68 (0.49 to 1.59) | |
| LTA (rs909253) | ||
| TT | 33 (41.8%) | 54 (40.9%) |
| CT | 35 (44.3%) | 56 (42.4%) |
| CC | 11 (13.9%) | 22 (16.7%) |
| χ2 (p value) CC+CT vs TT | 0.02 (0.90) | |
| OR (95% CI) | 0.97 (0.55 to 1.70) | |
| LTA (rs1041981) | ||
| CC | 33 (41.8%) | 48 (36.4%) |
| AC | 33 (41.8%) | 71 (53.8%) |
| AA | 13 (16.4%) | 13 (9.8%) |
| χ2 (p value) AA+AC vs CC | 0.61 (0.43) | |
| OR (95% CI) | 0.80 (0.45 to 1.41) | |
| TNF (rs1799724) | ||
| CC | 40 (50.6%) | 88 (66.7%) |
| CT | 21 (26.6%) | 30 (22.7%) |
| TT | 18 (22.8%) | 14 (10.6%) |
| χ2 (p value) TT+CT vs CC | 5.32 (0.021) | |
| OR (95% CI) | 1.95 (1.10 to 3.45) | |
| LST1 (rs2256965) | ||
| CC | 23 (29.1%) | 52 (39.4%) |
| CT | 38 (48.1%) | 57 (43.2%) |
| TT | 18 (22.8%) | 23 (17.4%) |
| χ2 (p value) TT+CT vs CC | 2.28 (0.13) | |
| OR (95% CI) | 1.58 (0.87 to 2.88) | |
| NCR3 (rs2736191) | ||
| GG | 5 (6.3%) | 18 (13.6%) |
| CG | 36 (45.6%) | 50 (37.9%) |
| CC | 38 (48.1%) | 64 (48.5%) |
| χ2 (p value) CC+CG vs GG | 2.28 (0.13) | |
| OR (95% CI) | 1.58 (0.87 to 2.88) | |
| AIF1 (rs2857697) | ||
| AA | 23 (29.1%) | 33 (25.0%) |
| AG | 38 (48.1%) | 67 (50.8%) |
| GG | 18 (22.8%) | 32 (24.2%) |
| χ2 (p value) GG+AG vs AA | 0.43 (0.51) | |
| OR (95% CI) | 0.81 (0.43 to 1.52) | |
| BAT2 (rs2242660) | ||
| TT | 14 (17.7%) | 28 (21.2%) |
| CT | 33 (41.8%) | 55 (41.7%) |
| CC | 32 (40.5%) | 49 (37.1%) |
| χ2 (p value) CC+CT vs TT | 0.38 (0.54) | |
| OR (95% CI) | 1.25 (0.61 to 2.55) | |
| BAT3 (rs3117583) | ||
| TT | 70 (88.6%) | 104 (78.8%) |
| CT | 8 (10.1%) | 27 (20.5%) |
| CC | 1 (1.3%) | 1 (0.7%) |
| χ2 (p value) CC+CT vs TT | 3.30 (0.069) | |
| OR (95% CI) | 0.48 (0.21 to 1.11) | |
Significant p values are in bold.
Table 3 haplotype block and haplotype‐based association analysis in the genomic region covering the TNFα locus.
| Haplotype block | Haplotype frequency | χ2 | p Value | ||
|---|---|---|---|---|---|
| Haplotype | Sequence | AS | Control | ||
| A | ATCACCG | 0.18 | 0.21 | 0.28 | 0.60 |
| B | GATCT*TC | 0.22 | 0.12 | 6.59 | 0.010 |
| C | AATCCCC | 0.09 | 0.11 | 0.55 | 0.46 |
| D | AATCCTC | 0.09 | 0.08 | 0.41 | 0.52 |
| E | ATCACCC | 0.06 | 0.09 | 0.13 | 0.71 |
| F | GATCCTC | 0.07 | 0.08 | 0.87 | 0.35 |
| G | AATCTTC | 0.04 | 0.03 | 0.36 | 0.55 |
*–850T Allele.
Haplotypes A–G include seven SNPs spanning a 58‐kb region.
Significant p values are in bold.
Genotype frequencies of all SNPs except SNP rs1799724 in control subjects did not deviate from Hardy–Weinberg equilibrium, and all SNPs except rs1799724 (χ2 = 5.324 p = 0.021, OR = 1.950, CI: 1.10–3.45) had no association with AS.
One haplotype block, in which we identified several common haplotypes including more than 70% of the individuals within both groups, comprised seven SNPs covering from BAT1 to NCR3. Haplotype‐based association analysis between case and control groups found that a haplotype including the –850T allele was strongly associated with AS (χ2 = 6.59, p = 0.01).
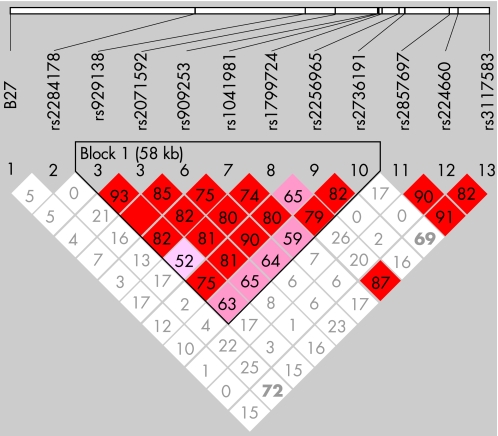
To determine linkage between the HLA‐B27 and TNFα genes, we selected a tag SNP on the HCP5 gene, from which the physical distance to these two genes was equal. We performed an LD test in both case and control groups and found that D′ between rs2284178 and the −850SNP was very low (0.012 in both groups) (fig 2).
Figure 2 Haplotype block and LD test in 100 kb genomic region covering TNFα gene are generated by Haploview V.3.32 software. Each box represents LD (range from 0 to 1) between pairs of SNPs. Red, strong LD; grey, high D′/low LOD value; white, low D′.
Discussion
To reconfirm the association of the TNFα locus with predisposition to AS, we typed 10 common SNPs in a 100 kb genomic region around the TNFα gene covering nine gene loci (BAT1, NFKBIL1, LTA, TNFα, LST1, NCR3, AIF1, BAT2 and BAT3), encoding molecules relevant to inflammation or autoimmune disorders. We found one block of intense LD in which D′ gradually reduced from the middle of the block towards BAT1 and NCR3 at the boundaries (fig 2) and found a haplotype that was significantly associated with AS by haplotype block analysis (table 3). Therefore we concluded that the gene loci for AS susceptibility are located between the two loci. The haplotype contained seven genes (BAT1, NFKBIL‐1, LTA, TNFα, LTB, LST1, NCR3) of which only the TNFα gene seemed to have supporting evidence to indicate it as a candidate gene for AS.
In this study, single allele‐based association analysis indicated that of the 10 SNPs selected from 9 working genes, only the TNFα –850C→T SNP was significantly associated with AS, which was in concordance with our previously studies performed in two other populations in China. The evidence from the LD test and single allele‐based association analysis forcefully suggested that the –850T allele confers susceptibility to AS. We evaluated the significance level by four methods, including the Bonferroni correction (see supplementary file 2; available at http://jmg.bmj.com/supplemental). Nearly all analysis results supported our study and the evidence from biological studies of TNFα also support the notion that this SNP variant is associated with AS.
The studies of the TNFα promoter function showed that the –850C nucleotide is involved in a consensus binding sequence (CCCCC*CTTAACGAAG) of the transcription factor nuclear factor (NF)‐κB p50/p50. The p50/p50 homodimers are capable of downregulating TNFα expression by acting as a repressor that inhibits the trans‐activating effects of NF‐κB p65 as they bind the DNA sequence.10,11 A C nucleotide on this sequence (denoted by the asterisk in the above sequence) substituted by A will specifically reduce inhibition of TNFα expression by the p50/p50 homodimer, in contrast to the enhancement of inducible TNF production in macrophages.12 However, whether the SNP –850C→T (underlined in the above sequence) is also involved in the mechanism implemented by the NF‐κB subunit complex through its influence on TNF gene expression remains to be confirmed.
Recently, one study showed that the –850 C nucleotide was also implicated in a conservative binding site (TAACG)13 of the transcription regulation factors V‐Myb and C‐Myb, which act in synergy with members of the C/EBP family to regulate the expression of myeloid‐specific genes. The c‐myb proto‐oncogene and the v‐Myb protein, a mutated form of the c‐Myb protein, are predominantly expressed in and induce the development of immature haematopoietic cells, and can result in monocytic leukaemia. However, it is as yet unknown whether the mechanism of upregulation of TNFα in the blood and synovial fluid of AS patients is correlated with V‐myb/C‐Myb production.
Combining our study data, we propose a hypothesis that the –850C→T variant in the binding region of the NF‐κB subunit complex/Myb may affect the transcription activation of the TNFα promoter by altering the interaction between transcription factors and DNA sequence. Previous studies have shown that the HLA‐B27 gene strongly predisposes to AS, but more recent studies have provided some evidence that non‐HLA‐B27 genetic factors also contribute to susceptibility to AS.14,15,16,17 In the current study, to further determine whether HLA‐B27 is linked to the TNFα gene, we selected a tag SNP (rs2284178)in the HCP5 gene, from which the physical distance was equal to these two genes. LD test showed that the D′ value (0.02) strongly indicated recombination between the two loci. In addition, we performed a stratification analysis based on HLA‐B27 negativity in case–case or case–control studies, and the result suggested a trend towards TNFα –850 association with AS. The carrier rate of the TNFα –850 mutant in HLA‐B27‐negative cases (62.5%) was higher than that in HLA‐B27‐7‐positive cases (47.9%). The carrier rate of the TNFα –850 mutant was higher in HLA‐B27 negative cases (62.5%) group than in the control group (31.7%). This suggested that the risk of AS is meaningful only for people with the TNFα –850 mutation and carriers negative for HLA‐B27 (supplementary tables 2–4; available at http://ard.bmj.com/supplemental). Therefore, the –850T allele may increase the risk of AS independently of the HLA‐B27 gene. However, it is still necessary to improve the control population construction of the Hardy–Weinberg equilbrium as a representative data item of population (supplementary file 1; available at http://jmg.bmj.com/supplemental).
Whether the TNFα gene is a genuine susceptibility gene for AS remains to be confirmed in other larger populations and the mechanism of action of the –850T variation on the entire disease process remains to be elucidated further.
In conclusion, this study provides an evidence in support of association of non‐HLA‐B27 genes with AS. The loci of genes predisposing to AS are limited to a 58 kb region around the TNFα gene, and the TNFα 850 C→T SNP is associated with susceptibility to AS independently of the HLA‐B27 gene.
Supplementary material is available on the ARD website at http://ard.bmj.com/supplemental
Copyright © 2007 BMJ Publishing Group and European League Against Rheumatism
Supplementary Material
Acknowledgements
We thank Professor Qingyu Zeng and Dr Zhiduo Hou, Department of Rheumatology, Shantou University Medical College; Dr Gang Wang, Huizhou Blood Center, Huizhou; and Dr Jianjun Yang from the Public Health School, Ningxia Medical College for supplying the blood samples from the patients with AS and healthy controls.
Abbreviations
AS - ankylosing spondylitis
HLA - human leucocyte antigen
LD - linkage disequilibrium
NF - nuclear factor
RG - restriction site‐generating
SNP - single‐nucleotide polymorphism
TNF - tumour necrosis factor
Footnotes
Funding: The study is supported partly by National Scientific Foundation of China (30471926, 30671110) and National Basic Research Program of China (973 Program) (2006CB503901) grants to Z Y.
Supplementary material is available on the ARD website at http://ard.bmj.com/supplemental
References
- 1.Davis J C, Jr, Van Der Heijde D, Braun J, Dougados M.et al Recombinant human tumor necrosis factor receptor (etanercept) for treating ankylosing spondylitis: a randomized, controlled trial. Arthritis Rheum 2003483230–3236. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J J, Baron G, Van Der Heijde D, Felson D T, Dougados M. Ankylosing spondylitis assessment group preliminary definition of short‐term improvement in ankylosing spondylitis. Arthritis Rheum 2001441876–1886. [DOI] [PubMed] [Google Scholar]
- 3.Braun J, Pham T, Sieper J, Davis J, van der Linden S J, Dougados J M.et al International ASAS consensus statement for the use of anti‐tumour necrosis factor agents in patients with ankylosing spondylitis. Ann Rheum Dis 200362817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu X Q, Zeng Q Y, Sun L, Wang G, Tang L, Hou Z D.et al Recognition and study of susceptible gene to ankylosing spondylitis. Hereditas 2005271–6. [PubMed] [Google Scholar]
- 5.van der Linden S M, Valkenburg H A, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum 198427361–368. [DOI] [PubMed] [Google Scholar]
- 6.Okamoto K, Makino S, Yoshikawa Y, Takaki A, Nagatsuka Y, Ota M.et al Identification of I kappa BL as the second major histocompatibility complex‐linked susceptibility locus for rheumatoid arthritis. Am J Hum Genet 200372303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T.et al Functional SNPs in the lymphotoxin‐alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 200232650–654. [DOI] [PubMed] [Google Scholar]
- 8.Steffens‐Nakken H M, Zwart G, van den Bergh F A J T M. Validation of allele‐specific polymerase chain reaction for DNA typing of HLA‐B27. Clin Chem 199541687–692. [PubMed] [Google Scholar]
- 9.Barrett J C, Fry B, Maller J, Daly M J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 200521263–265. [DOI] [PubMed] [Google Scholar]
- 10.Prkins N D, Schmid R M, Duckett C S, Leung K, Rice N R, Nabel G J. Distinct combinations of NF‐κB subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci U S A 1992891529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz M L, Baeuerle P A. The p65 subunit is responsible for the strong transcription activating potential of NF‐κB. EMBO J 1991103805–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udalova I A, Richardson A, Denys A, Smith S, Ackerman H.et al Functional consequences of a polymorphism affecting NF‐κB p50‐p50 binding to the TNF promoter region. Mol Cell Biol 2000209113–9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weston K. Extension of the DNA binding consensus of the chicken c‐Myb and v‐Myb proteins. Nucleic Acids Res 1992203043–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin A, Marder A, Becks E, Burns T. Genetic differences between B27 positive patients with ankylosing spondylitis and B27 positive healthy controls. Arthritis Rheum 1983261460–1464. [DOI] [PubMed] [Google Scholar]
- 15.Laval S H, Timms A, Edwards S, Bradbury L, Brophy S, Milicic A.et al Whole‐genome screening in ankylosing spondylitis: evidence of non‐MHC genetic‐susceptibility loci. Am J Hum Genet 200168918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou C T, Timms A E, Wei J C, Tsai W C, Wordsworth B P, Brown M A. Replication of association of IL1 gene complex members with ankylosing spondylitis in Taiwanese Chinese. Ann Rheum Dis 2006651106–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timms A E, Crane A M, Sims A M, Cordell H J, Bradbury L A, Abbott A.et al Related Articles, Links The interleukin 1 gene cluster contains a major susceptibility locus for ankylosing spondylitis. Am J Hum Genet 200475587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.