Abstract
Background
Certain sequences present in the hypervariable region of human leucocyte antigen (HLA)‐DRB1 known as the shared epitope (SE) are hypothesised to increase the risk of rheumatoid arthritis (RA), whereas alleles encoding aspartic acid at position 70 (D70 alleles) may have a protective effect.
Methods
Patient HLA‐DRB1 serotypes were assessed and the genotypes encoding the SE motif or the putatively protective D70 motif identified in a large RA cohort. Logistic regression was used to analyse associations of genotype with presence of disease, comorbidities and disease severity, and association between genotype and change in disease activity over time.
Results
The 689 patients enrolled had a mean (SD) age of 57.9 (13.7) years and mean (SD) disease duration of 15.3 (12.7) years. In a comparison with 482 ethnicity matched population‐based controls, the D70 sequence exerted a strong protective effect (OR = 0.52, p<0.001) that remained significant when the SE at the same locus was accounted for (OR = 0.72, 95% CI 0.60 to 0.86, p<0.001). The SE assessed on all HLA‐DRB1 serotypic backgrounds except DR1 was associated with RA susceptibility (additive OR = 2.43, p<0.001). Associations were found between SE and serum levels of rheumatoid factor (p<0.001, with correlation of 0.18) and anti‐cyclic citrullinated peptide antibodies (p<0.001, with correlation of 0.25) but not with serum C‐reactive protein.
Conclusion
The D70 allele has a significant protective effect that is mitigated but still significant when the risk effect of the SE at the same locus is taken into account. The presence of the SE on DR4 is associated with greater RA susceptibility and certain disease‐activity measures.
Keywords: rheumatoid arthritis, genetics, shared epitope, patient registry
Recent research has emphasised the importance of early treatment for the chronic inflammatory illness rheumatoid arthritis (RA). As RA drugs become more targeted and effective with the introduction of the newer biologicals, the ability to identify and classify an individual's disease by severity, key manifestations and progression rate grows ever more important for optimal care. However, despite impressive advances in treatment of RA over the past 20 years, nearly 30% of patients do not achieve an adequate clinical response to treatment as defined by American College of Rheumatology (ACR) response criteria.1,2,3,4
Genetic analysis offers a unique opportunity to rapidly and accurately assess a patient's disease profile through the determination of alleles related to disease severity, changes in activity or functional status, and potentially even efficacy of certain drugs. The Brigham and Women's Rheumatoid Arthritis Sequential Study (BRASS) is a study collecting prospective genetic, proteomic and clinical data on >900 patients with RA in the Robert Breck Brigham Arthritis Center. We studied a large cohort of patients with RA to evaluate genetic associations in the context of other disease markers, comorbidities and general disease progression.
Recent studies have determined that the regions of the human leucocyte antigen (HLA) DRB1 associated with RA share a set of polymorphisms in the amino acid sequence from residues 67 to 74, termed the shared epitope (SE).5 Present on most DR4, DR10, DR14 and some DR1 alleles,5,6 the SE sequence varies by only a few residues across the major DR4 and DR1 subtypes.7,8 The process by which the SE confers disease risk is not fully understood.9 It lies within the allelic hypervariable regions (AHVR) that encode the antigen‐binding site and is therefore likely to affect the array of peptides bound and presented to CD4+ T lymphocytes via the T‐cell receptor.7,10,11
In the past 5 years, cohort studies have identified another polymorphism in the hypervariable region homologous across certain HLA alleles. At position 70 in the AHVR, there is an amino acid substitution of aspartic acid (D) in lieu of glutamine (Q) or arginine (R), which has been suggested to reduce RA risk, perhaps through a similar mechanism to the SE.8,12,13,14 Little has been reported regarding association of the D70 allele with aspects of RA beyond decreased susceptibility to disease. Some have speculated that presence of the D70 should correlate with fewer radiographic changes and perhaps lower levels of inflammatory biomarkers. For both the D70 and SE alleles, there is little consensus, and increasingly, conflicting data regarding their relevance to disease progression and activity.
In this study, we set out to evaluate associations between HLA‐DRB1 alleles, particularly the SE and D70 regions, which have previously reported in the literature, to determine the strength of the risk or protective effect conferred. We then explored associations between these regions and certain clinical and blood‐based markers of disease severity. Finally, we looked at changes in inflammatory markers and disease status over time to determine whether these alleles are associated with more rapid positive or negative changes in disease state.
Methods
Patient recruitment
All study protocols were approved by the Brigham and Women's Hospital internal review board. We recruited patients seeing rheumatologists at the Robert Breck Brigham Arthritis Center at Brigham and Women's Hospital. Patients were eligible to join the study if they had a diagnosis of RA, were >18 years of age and did not have a diagnosis of psoriatic arthritis or systemic lupus erythematosus. We initially screened for participants based on billing diagnosis (714.0 for RA or 714.3 for seronegative inflammatory arthritis) and then verified the diagnosis and other conditions with the rheumatologist. Eligible patients received a letter inviting them to join the study at their next appointment, at which they were approached by their rheumatologist and a research assistant and enrolled if they agreed to join. Only 10% of the subjects approached declined participation.
Study procedures
Patients received yearly study examinations at the clinic, including blood specimens, questionnaires and joint examinations, and completed a self‐administered postal questionnaire at 6‐month intervals between clinic visits. At enrolment, each participant completed a detailed interview and self‐administered questionnaires providing demographic and clinical information, and potential risk factors such as smoking history, arthritis and non‐arthritis drugs, comorbidities and surgeries. Assessment of functional status was performed using the self‐efficacy survey and the Multidimensional Health Assessment Questionnaire (MDHAQ) and evaluation of disease activity using pain, fatigue and functional ability scales.15,16,17 Each participant received an examination by a doctor to assess joint swelling and pain, extra‐articular features and general disease activity using a visual analogue scale. The joint examination included a standard 28‐joint count according to ACR response criteria and inspection of joints for swelling and tenderness, providing an assessment of disease activity (ACR core set).17 Patients also received hand and wrist radiographs and provided samples of blood for standard laboratory panel, including C‐reactive protein (CRP), anti‐CCP antibody titre and rheumatoid factor (RF), and for DNA analysis.
Every subsequent year, using interviews and self‐administered questionnaires, patients provided updated information on demographics, drugs and comorbidities, and additional information about pregnancies, hospitalisations, exercise regimens and social support. Additionally, they completed scales assessing depression, functional ability, fatigue and pain and provided additional blood for serum samples. Doctors completed joint assessments and overall health assessments. Every 2 years, patients received additional hand and wrist radiography.
At 6‐month intervals between the yearly visits, patients completed self‐administered questionnaires with information about functional status, drug use, general health state and resource use.
HLA genotyping
We extracted DNA from whole blood using a commercially available kit (Qiagen, Valencia, California, USA). HLA‐DRB1 serotypes were assessed from DNA sequences using allele‐specific PCR methods based on published PCR primers and HLA‐DRB1 intron sequences.18 The primer pairs we used identify DNA sequences corresponding to serotypes DR1, DR3, DR4, DR7, DR8, DR9, DR10, DR11+DR13, DR12, DR14, DR15 and DR16. In addition to serotype, we sought to identify which patient genotypes encoded either the SE motif (QKRAA, QRRAA or RRRAA at positions 70–74)19or the putatively protective motif12 encoding an aspartic acid residue at position 70 (D70 alleles). PCR products for alleles encoding a serotype that included some SE and/or D70 motifs (DR1, DR4, DR14) were sequenced using standard dye‐terminator chemistry (Applied Biosystems, Foster City, California, USA) to determine which motifs each patient's genotype contained. Other serotypes effectively encode only one type of motif (SE (DR10) or D70 (DR7, DR8, DR11, DR12, DR13, DR16)) or neither (DR3, DR9, DR15); these alleles were not sequenced (see the IMGT HLA Sequence Database at http://www.ebi.ac.uk/imgt/hla/index.html for detailed information on distribution of sequence motifs across classical serotypes). The genotyping was only uncertain in two cases and these were deleted from the analysis pool. To avoid confounding of differences in serotype frequencies across ethnic groups with association of serotype and disease risk, we only typed subjects reporting Caucasian ethnicity.
For analysis of HLA allelic association with RA disease risk, a healthy control population was required. This population was selected by random digit dialling from the Baltimore area of Maryland (Innovative Medical Research Inc., now a subsidiary of Caremark Inc. performed the ascertainment). The random digit dialling procedure using a population‐sampling design recruited a subject pool that is a random subset of the general population. All subjects provided broad informed consent for use of their DNA in disease research. They all additionally identified themselves as being of European Caucasian ancestry and provided extensive medical history information via questionnaire. Patients indicating a history of RA were not used for this study.
Statistical analysis
We used logistic regression to evaluate the association of HLA‐DRB1 genotype with categorical variables including case/control status, presence of rheumatoid nodules, comorbid Sjogren syndrome, radiographic changes (erosion or periarticular osteopenia), morning stiffness and requirement for joint‐replacement surgery (JRS). Association of genotype with continuous traits was tested using general linear regression or Wilcoxon rank sum test if applicable. We investigated HLA‐DRB1 influence on serum levels of CRP, RF and anti‐CCP antibodies, as well as patient and physician ratings of disease activity, and age at onset of disease. Variables that were not normally distributed were transformed to achieve the criteria for linear regression analysis.
Results
Cohort clinical description
The participants were, on average, in their fifth decade and mostly women (81%). The group had longstanding RA at baseline (mean (SD) disease duration 15.3 (12.7 years). The doctors reported that 62% of patients were seropositive and 38% had rheumatoid nodules. Most had active disease at recruitment with a median CRP level of 3.4 mg/l, and morning stiffness duration >1 hour for 34% of the cohort. The mean Disease Activity Score for CRP (DAS28‐CRP) was 3.4 (1.4), indicating moderate disease activity. The doctors reported a median of six swollen joints and seven painful joints. Most patients indicated a mild level of functional impairment with a median MDHAQ score of 0.6. Table 1 describes the clinical characteristics of the population.
Table 1 Clinical characteristics of the BRASS cohort .
| Clinical variable | Value |
|---|---|
| Age, mean (SD) | 57.9 (14) |
| Female gender, n (%) | 561 (81) |
| Duration of disease (years), mean (SD) | 15.3 (12.7) |
| Age at diagnosis, mean (SD) | 42.6 (15.1) |
| Symptom duration before diagnosis (years)* | 0 (0 to 2) |
| DAS (0–10), mean (SD) | 3.4 (1.4) |
| C‐reactive protein (mg/l)* | 3.4 (1.3 to 9.6) |
| Rheumatoid factor >15 mg/dl, n (%) | 420 (61.9) |
| Presence of nodules, n (%) | 255 (37.9) |
| Total swollen joint count* | 6 (2 to 13) |
| Total painful joint count* | 7 (1 to 14) |
| Morning stiffness >1 hour, n (%) | 157 (33.6) |
| MDHAQ (0–3)* | 0.6 (0.2 to 1) |
| Physician global assessment (0–10)* | 3 (2 to 3) |
| Patient global assessment (0–100)* | 25 (10 to 50) |
| >3 Flares in the previous 6 months, n (%) | 150 (33.5) |
BRASS, Brigham and Women's Rheumatoid Arthritis Sequential Study; DAS, Disease Activity Score; MDHAQ, Multidimensional Health Assessment Questionnaire.
*Values are median (25th to 75th centile).
Cohort genetic profile
HLA‐DRB1 association with disease risk
HLA‐DRB1 serotypes were determined for a total of 689 subjects from the BRASS study and 482 controls. Serotype frequencies and frequency of SE and D70 sequences within each serotype are shown in table 2. The common RA serotype, DR4, appeared more often in BRASS subjects (34.8%) than in controls (17.4%), whereas the other common RA serotype, DR1, which bears the SE, was equally prevalent in both groups (13.1% BRASS, 11.8% controls). We also observed that the SE was more common on DR4 alleles in patients with RA than in controls (89.4% vs 82.1%, p = 0.022); this was not the case for SE sequences on DR1 alleles (93.3% vs 89.5%, p = 0.24).
Table 2 HLA‐DRB1 serotype frequencies observed in patients with RA and control subjects .
| Serotype | BRASS | Control | ||||
|---|---|---|---|---|---|---|
| n (%) | SE, n (%) | D70, n (%) | n (%) | SE, n (%) | D70, n (%) | |
| DR1 | 180 (13.1) | 168 (93.3) | 12 (6.7) | 114 (11.8) | 102 (89.5) | 12 (10.5) |
| DR2 (15) | 148 (10.7) | 119 (12.3) | ||||
| DR2 (16) | 22 (1.6) | 22 (100) | 12 (1.2) | 12 (100) | ||
| DR3 | 147 (10.7) | 121 (12.6) | ||||
| DR4 | 479 (34.8) | 428 (89.4) | 24 (5) | 168 (17.4) | 138 (82.1) | 10 (6) |
| DR7 | 100 (7.3) | 100 (100) | 127 (13.2) | 127 (100) | ||
| DR8 | 25 (1.8) | 25 (100) | 22 (2.3) | 22 (100) | ||
| DR9 | 12 (0.9) | 5 (0.5) | ||||
| DR10 | 23 (1.7) | 23 (100) | 7 (0.7) | 7 (100) | ||
| DR11/DR13 | 214 (15.5) | 214 (100) | 241 (25) | 241 (100) | ||
| DR12 | 6 (0.4) | 6 (100) | 3 (0.3) | 3 (100) | ||
| DR14 | 22 (1.6) | 2 (9.1) | 25 (2.6) | 1 (4) | ||
| Totals | 1378 | 621 (45.1) | 403 (29.2) | 964 | 248 (25.7) | 427 (44.3) |
BRASS, Brigham and Women's Rheumatoid Arthritis Sequential Study; HLA, human leucocyte antigen; RA, rheumatoid arthritis; SE, shared eipitope.
Based on these observations, we chose to test a series of four disease‐risk association models: DR4, DR4‐SE, DR1‐SE and non‐DR1‐SE (table 3). All models except for the DR1‐SE background showed robust association with RA risk. The strongest association (additive OR = 2.43, 95% CI 1.98 to 2.97) was observed for non‐DR1‐SE. We therefore used this coding of HLA‐DRB1 genotype (non‐DR1‐SE) for all subsequent analyses. For purposes of brevity, these alleles are referred to hereafter simply as SE alleles.
Table 3 Odds ratios for four disease‐risk association models and the hypothesised protective D70 allele.
| Risk factor | OR | 95% CI | p Value |
|---|---|---|---|
| SE | 2.09 | 1.77 to 2.48 | <0.001 |
| DR4 | 2.29 | 1.88 to 2.78 | <0.001 |
| Non DR1‐SE | 2.43 | 1.98 to 2.97 | <0.001 |
| DR1‐SE | 1.15 | 0.89 to 1.47 | NS |
| D70 | 0.72 | 0.60 to 0.86 | <0.001 |
NS, non‐significant; SE, shared epitope.
*No homozygosity effect found.
All models tested were additive, so ORs represent differential RA risk per copy of the evaluated allele.
D70 analysis was controlled for presence of the shared epitope.
When considered in isolation, the D70 sequence (present on many different serotypic backgrounds; table 2) exerted a strong protective effect (OR = 0.52, p <0.001), reducing RA risk, as hypothesised by Mattey et al and subsequently asserted by others.12,20,21 When the presence of the risk‐associated SE at the same locus was taken into account, the independent effect of D70 was more modest in size, but remained statistically significant (table 3). No interaction between HLA‐DRB1 genotype and gender was detected in these analyses. Although our cohort has a much larger percentage of females than males, the SE and D70 alleles modified risk to a comparable degree for both.
HLA‐DRB1 association with disease phenotype
We evaluated SE association with five disease phenotypes that commonly occur in RA, including Sjogren syndrome, morning stiffness, rheumatoid nodules, radiographically visible changes in any joint and the need for JRS. All analyses were controlled for age, gender and duration of RA. Possession of one or two SE alleles was additively associated with a greater likelihood of JRS, rheumatoid nodules and radiographic changes, but had no influence on the likelihood of a patient developing Sjogren syndrome or morning stiffness (table 4). We observed a modestly significant interaction between gender and SE in association with JRS (p = 0.045); the SE appeared to be a more potent risk factor for men than for women.
Table 4 Odds ratios for HLA‐DRB1 SE association with five RA‐related disease phenotypes, controlled for age, gender, and duration of RA.
| Phenotype | OR | 95% CI | p Value |
|---|---|---|---|
| Comorbid Sjogren | 0.98 | 0.79 to 1.21 | NS |
| Joint replacement | 1.47 | 1.08 to 2.00 | 0.016 |
| Morning stiffness | 0.94 | 0.74 to 1.21 | NS |
| Rheumatoid nodules | 1.71 | 1.35 to 2.16 | <0.001 |
| Radiographic change | 1.54 | 1.13 to 2.11 | <0.001 |
HLA, human leucocyte antigen; NS, non‐significant; RA, rheumatoid arthritis; SE, shared epitope.
All ORs were calculated for an additive genetic model, and so reflect increased risk of exhibiting the phenotype per additional SE allele.
Radiographic change denotes documented bone erosion or periarticular osteopenia in any joint.
HLA‐DRB1 association with disease activity
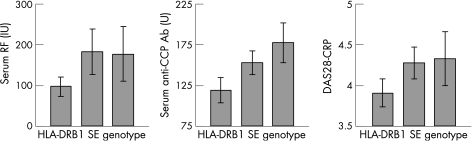
We also tested association of SE with several continuous parameters descriptive of or often associated with RA disease activity: rheumatoid factor, CRP, anti‐CCP antibodies, DAS28‐CRP and patient's global assessment of disease activity. Analyses were again controlled for age, gender and duration of disease. We observed a significant association with serum rheumatoid factor (p<0.001). This association did not change after the model was controlled for smoking status, although smoking was also significantly associated with serum RF (p<0.001), but not with serum CRP (fig 1). Presence of SE alleles interacted significantly with gender in association with serum RF (p = 0.004); a much stronger positive association between number of SE alleles and increasing RF was observed in males. Most patients were positive for anti‐CCP antibodies (66.5%), and positivity was significantly associated with the SE. This association held after adjustment for smoking status (p<0.001); 49.8% of patients with no SE alleles were positive, versus 75.3% of patients with at least one SE allele (p<0.001). Among anti‐CCP antibody‐positive patients, there was an additional, strong association between serum antibody level and SE genotype (p<0.001).
Figure 1 Association of HLA‐DRB1 SE with two serum proteins characteristic of RA, and with the composite RA disease activity metric DAS28‐CRP.
The SE was associated with greater disease activity as measured by DAS28‐CRP (p = 0.013) but not with patient's global assessment of disease activity. We observed a modest association between presence of SE alleles and earlier age at disease onset (p = 0.019, each SE allele showed an age of onset 1.7 years earlier). The D70 allele was not found to associate with any tested aspect of disease phenotype.
Finally, we tested both the SE and the D70 for association with change in disease activity as measured by anti‐CCP, DAS28‐CRP, swollen and painful joint counts and change in functional status as measured by the MDHAQ. We found no significant associations with change in any of these measures from baseline to 1 year with the presence of the SE or D70 (results not shown).
Discussion
In our cohort of 689 patients of Caucasian ancestry with RA, the presence of the D70 sequence on multiple DRB1 haplotypic backgrounds appeared to exert a significant protective effect against RA. The effect remained significant even when the increased risk associated with the SE at the same locus was factored into the analysis. Before accounting for the presence of the SE, the proportion of patients with RA carrying at least one D70 allele was substantially lower than that of controls (OR = 0.56, p<0.001). This risk ratio is similar in magnitude to those reported initially by Mattey et al for those with two D70 alleles only, in populations from the UK and Spain respectively (OR = 0.23 and 0.34, respectively), and by Ruiz‐Morales et al in a Mexican Mestizo population (OR = 0.4, 95% CI 0.2 to 0.7, p = 0.004).12,21 Mattey et al found that individuals carrying one allele positive for the SE and one D70 allele had no increased risk of disease.12 These earlier studies, however, did not take into account the presence of the SE at the same locus, and analysed the effect on RA risk of the D70 low‐risk allele at HLA‐DRB1 without accounting for unequal allele and genotype frequency expectations in cases and controls.12,20,21 Controlling for the increased risk contributed by the presence of an SE allele reduces but does not eliminate the apparent protective effect of the D70 allele (OR = 0.72, 95% CI 0.60 to 0.86, p<0.001). Because patients with RA are more likely to carry one or two SE alleles, they are expected, on that basis alone, to exhibit lower frequencies of all other possible genotypes, including homozygous D70 genotypes, than are healthy controls. Previous studies typically attempted to exclude this bias in comparing the frequency of D70/D70 genotypes in patients with RA and controls—for example by investigating subjects with no SE alleles, regardless of disease status. As described above, such studies reported strong protective effects, with ORs ranging from 0.19 to 0.40. To correctly estimate the independent protective effect of D70, however, it is necessary to use logistic regression to model the effect of SE and D70 alleles on disease risk simultaneously, a method that enables us to arrive at an additive OR of 0.72 (table 3).
In concordance with previous studies, we found no associations between D70 alleles and any aspect of disease phenotype, including comorbidities, disease activity and change in disease status over time.21 Mattey et al were unable to draw any conclusions regarding the effects of the D70 on radiographic progression or rheumatoid factor.12 In our cohort, the presence of the D70 allele had similar effects in men and women, and in those with different levels of disease duration, again in concordance with the results of previous studies.21
Since Gregersen's initial discovery of the third hypervariable domain, multiple studies, including his own analysis, have reported association of disease susceptibility with the presence of the SE on various HLA‐DRB1 serotypes, particularly DR1 and DR4.5,7,21 Our results indicate association with disease susceptibility only when the hypervariable region SE is present on alleles of non‐DR1 serotype, the majority of which are DR4 (additive OR = 2.43, 95% CI 1.98 to 2.97). These results are somewhat consistent with Anaya et al, who observed risk association in a Colombian population to be restricted to DR4 QRRAA SE sequences.22 Our estimate of the OR is similar to many published reports but lower than those reported in some studies.6,20,21 Variation in OR estimates among studies may be due to frequency differences in HLA alleles across populations. Similar to our findings with the D70 residue, we found no interaction between SE genotype and gender in predicting disease risk.
Although previous studies have shown the SE to be strongly implicated in RA risk, questions remain in two key areas: its predictive ability regarding the RA subtype and whether the SE offers any clues about disease progression. Multiple studies have reported the SE as a marker for more severe, and particularly more erosive disease, although this effect seems to vary across populations.23,24,25 Data from a large cross‐sectional cohort in England indicated that associations between SE genotype and radiographic severity are probably indirect and driven by an association with anti‐CCP antibodies.26 Data from some cohorts suggest a stronger effect in seronegative patients, whereas others have indicated correlations with certain extra‐articular manifestations.9,27,28 A large Swedish cohort recently reported an increased association with extra‐articular RA (OR = 1.79, 95% CI 1.04 to 3.08) and vasculitis (OR = 2.44, 95% CI 1.22 to 4.89).25 Other studies, however, have simply failed to find any association beyond disease susceptibility.29,30,31
In our large Caucasian cohort, we found that presence of the SE, in addition to imparting increased risk for RA, was associated with radiographic change and several common phenotypes. In addition to radiographic change (OR 1.54, 95% CI 1.13 to 2.11), joint replacement surgery (JRS) was associated with the SE (OR 1.47, 95% CI 1.08 to 2.00). Some previous studies have reported a similar correlation between DR4 alleles and erosive disease, as evidenced by radiographic changes,32,33,34,35 but others have reported none.29,36,37 Furthermore, our data indicate an apparently dominant mode of SE influence on disease activity as measured by DAS28‐CRP, with SE heterozygotes and homozygotes showing a similar increment in disease activity over those patients bearing no SE alleles (fig 1). In the absence of a direct association between SE and serum CRP in our study, this association presumably is driven by SE association with the other DAS28 components, tender and swollen joint counts. This observation may reflect the same underlying influence of SE on disease pathophysiology as the association between SE and radiographic changes in joint condition and requirement for JRS in our cohort (table 4).
Despite associations with greater disease activity and severity at study baseline, we observed no association of HLA‐DRB1 genotype with change in disease status from baseline to 1 year, as measured using a variety of validated functional measures and inflammatory markers.
Finally, in our cohort, there was not only a highly significant association between SE genotype and anti‐CCP positivity, but also, within anti‐CCP positive patients, a strong additive relationship between the number of SE alleles present and serum antibody titre (fig 1). Recently, Irigoyen et al described a significant association between anti‐CCP antibody positivity and the DRB1 SE for patients enrolled in the North American Rheumatoid Arthritis Consortium family cohort and the Study of New Onset Rheumatoid Arthritis,38 and Berglin et al and others saw no significant association with their subjects.8,39,40 Our results also confirmed several frequently reported associations; the presence of the SE is strongly associated with rheumatoid nodules, rheumatoid factor and younger age at disease onset. However, the implications for the mode of action of the allele of our observation of SE association with serum RF titre differed somewhat from that reported by Fries et al.6 That group found an association of SE homozygosity, but not heterozygosity, with RF positivity, suggesting a recessive mode of action. Our results suggest an entirely dominant role for the SE, with heterozygotes exhibiting a mean serum RF level equal to that observed in homozygotes (fig 1).
Limitations
Our study has several limitations with respect to the types of patients enrolled in BRASS, their disease durations and their drug histories. HLA alleles have been shown to confer distinct risks at different points of the illness. The varying disease durations of the patients in our cohort may have statistically affected our analysis of the susceptibility alleles. By not dividing the group into recent onset and longer disease duration, we may have diluted what would be a stronger effect in one population or the other. In addition, we do not show that the risk for SE on a DR1 background significantly differs from the risk encoded by SE on a DR4 background. The reduced risk encoded by SE on DR1 is likely to be due to the absence of correction for the much larger group of SE on DR4. Finally, the frequency of HLA alleles varies by ethnicity. Our study contained only Caucasian patients and thus our findings may be less applicable to other ethnic groups.
Conclusion
Our study of a large single‐centre RA cohort has shown a significant protective effect of the D70 allele at HLA‐DRB1, controlled for risk conferred by the SE at the same locus. We confirmed a significant risk associated with non‐DR1‐SE but did not observe any significant risk associated with DR1‐SE. We observed a number of associations between the SE and disease phenotypes but no associations between the D70 and any phenotypes or inflammatory markers. We observed an additive relationship of the number of SE alleles with anti‐CCP positivity, as well as associations with radiographic change and JRS and a dominant mode of SE influence on disease activity as measured by the DAS28‐CRP. We found no associations with change in disease status or disease activity over time for either allele. We suggest that prospective studies are warranted to determine the full clinical implications of our findings.
Acknowledgements
We would like to thank the study participants in the BRASS cohort and their rheumatologists who have donated their time to this work.
Abbreviations
ACR - American College of Rheumatology
AVHR - allelic hypervariable regions
BRASS - Brigham and Women's Rheumatoid Arthritis Sequential Study
CCP - cyclic citrullinated peptide
CRP - C‐reactive protein
DAS - Disease Activity Score
HLA - human leucocyte antigen
JRS - joint‐replacement surgery
MDHAQ - Multidimensional Health Assessment Questionnaire
RA - rheumatoid arthritis
RF - rheumatoid factor
SE - shared epitope
Footnotes
Millenium Pharmaceuticals, Inc funded establishment of the patient cohort used for these analyses.
References
- 1.Bridges S L., Jr The genetics of rheumatoid arthritis: influences on susceptibility, severity, and treatment response. Curr Rheumatol Rep 19991164–171. [DOI] [PubMed] [Google Scholar]
- 2.Moreland L. Unmet needs in rheumatoid arthritis. Arthritis Res Ther 20057(suppl 3)S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett A N, Peterson P, Zain A, Grumley J, Panayi G, Kirkham B. Adalimumab in clinical practice. Outcome in 70 rheumatoid arthritis patients, including comparison of patients with and without previous anti‐TNF exposure. Rheumatology (Oxford) 2005441026–1031. [DOI] [PubMed] [Google Scholar]
- 4.Weinblatt M E. Efficacy of methotrexate in rheumatoid arthritis. Br J Rheumatol 199534(Suppl 2)43–48. [PubMed] [Google Scholar]
- 5.Gregersen P K, Silver J, Winchester R J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 1987301205–1213. [DOI] [PubMed] [Google Scholar]
- 6.Fries J F, Wolfe F, Apple R, Erlich H, Bugawan T, Holmes T.et al HLA‐DRB1 genotype associations in 793 white patients from a rheumatoid arthritis inception cohort: frequency, severity, and treatment bias. Arthritis Rheum 2002462320–2329. [DOI] [PubMed] [Google Scholar]
- 7.Wordsworth B P, Lanchbury J S, Sakkas L I, Welsh K I, Panayi G S, Bell J I. HLA‐DR4 subtype frequencies in rheumatoid arthritis indicate that DRB1 is the major susceptibility locus within the HLA class II region. Proc Natl Acad Sci U S A 19898610049–10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries N, Tijssen H, van Riel P L, van de Putte L B. Reshaping the shared epitope hypothesis: HLA‐associated risk for rheumatoid arthritis is encoded by amino acid substitutions at positions 67–74 of the HLA‐DRB1 molecule. Arthritis Rheum 200246921–928. [DOI] [PubMed] [Google Scholar]
- 9.Weyand C M, McCarthy T G, Goronzy J J. Correlation between disease phenotype and genetic heterogeneity in rheumatoid arthritis. J Clin Invest 1995952120–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu X T, Bono C P, Woulfe S L, Swearingen C, Summers N L, Sinigaglia F.et al Pocket 4 of the HLA‐DR(alpha,beta 1*0401) molecule is a major determinant of T cells recognition of peptide. J Exp Med 1995181915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirschmann D A, Duffin K L, Smith C E, Welply J K, Howard S C, Schwartz B D.et al Naturally processed peptides from rheumatoid arthritis associated and non‐associated HLA‐DR alleles. J Immunol 19951555655–5662. [PubMed] [Google Scholar]
- 12.Mattey D L, Dawes P T, Gonzalez‐Gay M A, Garcia‐Porrua C, Thomson W, Hajeer A H.et al HLA‐DRB1 alleles encoding an aspartic acid at position 70 protect against development of rheumatoid arthritis. J Rheumatol 200128232–239. [PubMed] [Google Scholar]
- 13.del Rincon I, Escalante A. HLA‐DRB1 alleles associated with susceptibility or resistance to rheumatoid arthritis, articular deformities, and disability in Mexican Americans. Arthritis Rheum 1999421329–1338. [DOI] [PubMed] [Google Scholar]
- 14.Reviron D, Perdriger A, Toussirot E, Wendling D, Balandraud N, Guis S.et al Influence of shared epitope‐negative HLA‐DRB1 alleles on genetic susceptibility to rheumatoid arthritis. Arthritis Rheum 200144535–540. [DOI] [PubMed] [Google Scholar]
- 15.Pincus T, Swearingen C, Wolfe F. Toward a multidimensional Health Assessment Questionnaire (MDHAQ): assessment of advanced activities of daily living and psychological status in the patient‐friendly health assessment questionnaire format. Arthritis Rheum 1999422220–2230. [DOI] [PubMed] [Google Scholar]
- 16.Ware J E, Jr, Sherbourne C D. The MOS 36‐item short‐form health survey (SF‐36). I. Conceptual framework and item selection. Med Care 199230473–483. [PubMed] [Google Scholar]
- 17.Pincus T, Strand V, Koch G, Amara I, Crawford B, Wolfe F.et al An index of the three core data set patient questionnaire measures distinguishes efficacy of active treatment from that of placebo as effectively as the American College of Rheumatology 20% response criteria (ACR20) or the Disease Activity Score (DAS) in a rheumatoid arthritis clinical trial. Arthritis Rheum 200348625–630. [DOI] [PubMed] [Google Scholar]
- 18.Kotsch K, Wehling J, Blasczyk R. Sequencing of HLA class II genes based on the conserved diversity of the non‐coding regions: sequencing based typing of HLA‐DRB genes. Tissue Antigens 199953486–497. [DOI] [PubMed] [Google Scholar]
- 19.Lanchbury J S, Panayi G S. Genetics of RA: the HLA shared epitope hypothesis and its implications. Br J Rheumatol 199130(Suppl 2)6–9. [PubMed] [Google Scholar]
- 20.Khani‐Hanjani A, Lacaille D, Horne C, Chalmers A, Hoar D I, Balshaw R.et al Expression of QK/QR/RRRAA or DERAA motifs at the third hypervariable region of HLA‐DRB1 and disease severity in rheumatoid arthritis. J Rheumatol 2002291358–1365. [PubMed] [Google Scholar]
- 21.Ruiz‐Morales J A, Vargas‐Alarcon G, Flores‐Villanueva P O, Villarreal‐Garza C, Hernandez‐Pacheco C, Yamamoto‐Furusho J K.et al HLA‐DRB1 alleles encoding the “shared epitope” are associated with susceptibility to developing rheumatoid arthritis whereas HLA‐DRB1 alleles encoding an aspartic acid at position 70 of the beta‐chain are protective in Mexican Mestizos. Hum Immunol 200465262–269. [DOI] [PubMed] [Google Scholar]
- 22.Anaya J M, Correa P A, Mantilla R D, Arcos‐Burgos M. Rheumatoid arthritis association in Colombian population is restricted to HLA‐DRB1*04 QRRAA alleles. Genes Immun 2002356–58. [DOI] [PubMed] [Google Scholar]
- 23.van Zeben D, Hazes J M, Zwinderman A H, Cats A, Schreuder G M, D'Amaro J, Breedveld F C. Association of HLA‐DR4 with a more progressive disease course in patients with rheumatoid arthritis. Results of a followup study. Arthritis Rheum 199134822–830. [DOI] [PubMed] [Google Scholar]
- 24.Thomson W, Pepper L, Payton A, Carthy D, Scott D, Ollier W.et al Absence of an association between HLA‐DRB1*04 and rheumatoid arthritis in newly diagnosed cases from the community. Ann Rheum Dis 199352539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turesson C, Schaid D J, Weyand C M, Jacobsson L T, Goronzy J J, Petersson I F.et al The impact of HLA‐DRB1 genes on extra‐articular disease manifestations in rheumatoid arthritis. Arthritis Res Ther 20057R1386–R1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mewar D, Coote A, Moore D J, Marinou I, Keyworth J, Dickson M C.et al Independent associations of anti‐cyclic citrullinated peptide antibodies and rheumatoid factor with radiographic severity of rheumatoid arthritis. Arthritis Res Ther 20068R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Combe B, Dougados M, Goupille P, Cantagrel A, Eliaou J F, Sibilia J.et al Prognostic factors for radiographic damage in early rheumatoid arthritis: a multiparameter prospective study. Arthritis Rheum 2001441736–1743. [DOI] [PubMed] [Google Scholar]
- 28.MacGregor A, Ollier W, Thomson W, Jawaheer D, Silman A. HLA‐DRB1*0401/0404 genotype and rheumatoid arthritis: increased association in men, young age at onset, and disease severity. J Rheumatol 1995221032–1036. [PubMed] [Google Scholar]
- 29.Eberhardt K, Fex E, Johnson U, Wollheim F A. Associations of HLA‐DRB and ‐DQB genes with two and five year outcome in rheumatoid arthritis. Ann Rheum Dis 19965534–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suarez‐Almazor M E, Tao S, Moustarah F, Russell A S, Maksymowych W. HLA‐DR1, DR4, and DRB1 disease related subtypes in rheumatoid arthritis. Association with susceptibility but not severity in a city wide community based study. J Rheumatol 1995222027–2033. [PubMed] [Google Scholar]
- 31.Citera G, Padulo L A, Fernandez G, Lazaro M A, Rosemffet M G, Maldonado Cocco J A. Influence of HLA‐DR alleles on rheumatoid arthritis: susceptibility and severity in Argentine patients. J Rheumatol 2001281486–1491. [PubMed] [Google Scholar]
- 32.Young A, Jaraquemada D, Awad J, Festenstein H, Corbett M, Hay F C.et al Association of HLA‐DR4/Dw4 and DR2/Dw2 with radiologic changes in a prospective study of patients with rheumatoid arthritis. Preferential relationship with HLA‐Dw rather than HLA‐DR specificities. Arthritis Rheum 19842720–25. [DOI] [PubMed] [Google Scholar]
- 33.Olsen N J, Callahan L F, Brooks R H, Nance E P, Kaye J J, Stastny P.et al Associations of HLA‐DR4 with rheumatoid factor and radiographic severity in rheumatoid arthritis. Am J Med 198884257–264. [DOI] [PubMed] [Google Scholar]
- 34.Calin A, Elswood J, Klouda P T. Destructive arthritis, rheumatoid factor, and HLA‐DR4. Susceptibility versus severity, a case‐control study. Arthritis Rheum 1989321221–1225. [DOI] [PubMed] [Google Scholar]
- 35.McMahon M J, Hillarby M C, Clarkson R W, Hollis S, Grennan D M. Major histocompatibility complex variants and articular disease severity in rheumatoid arthritis. Br J Rheumatol 199332899–902. [DOI] [PubMed] [Google Scholar]
- 36.Valenzuela‐Castano A, Garcia‐Lopez A, Perez‐Vilches D, Rodriguez‐Perez R, Gonzalez‐Escribano M F, Nunez‐Roldan A. The predictive value of the HLA shared epitope for severity of radiological joint damage in patients with rheumatoid arthritis. A 10 year observational prospective study. J Rheumatol 200027571–574. [PubMed] [Google Scholar]
- 37.Rau R, Herborn G, Zueger S, Fenner H. The effect of HLA‐DRB1 genes, rheumatoid factor, and treatment on radiographic disease progression in rheumatoid arthritis over 6 years. J Rheumatol 2000272566–2575. [PubMed] [Google Scholar]
- 38.Welsing P M, Landewe R B, van Riel P L, Boers M, van Gestel A M, van der Linden S.et al The relationship between disease activity and radiologic progression in patients with rheumatoid arthritis: a longitudinal analysis. Arthritis Rheum 2004502082–2093. [DOI] [PubMed] [Google Scholar]
- 39.van Gaalen F A, van Aken J, Huizinga T W, Schreuder G M, Breedveld F C, Zanelli E.et al Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum 2004502113–2121. [DOI] [PubMed] [Google Scholar]
- 40.Berglin E, Padyukov L, Sundin U, Hallmans G, Stenlund H, Van Venrooij W J.et al A combination of autoantibodies–cyclic citrullinated peptide (CCP) and HLA‐DRB1 locus antigens is strongly associated with future onset of rheumatoid arthritis. Arthritis Res Ther 20046R303–R308. [DOI] [PMC free article] [PubMed] [Google Scholar]