Abstract
The retinoblastoma tumor suppressor protein (pRB) is a potent inhibitor of mammalian cell growth and the functional inactivation of pRB is widely presumed to be essential for progression of the cell cycle from G1 phase. In this work, the generality of pRB-based cell cycle control in mammalian cells was addressed by conditionally expressing pRB in cytokine-dependent hematopoietic cells. We show herein that these cells are able to progress through the cell cycle in response to cytokine despite the continued presence of supraphysiological amounts of wild-type pRB or phosphorylation-resistant pRB mutants. However, their growth was strongly blocked by ectopic expression of the pRB-related pocket protein, p130. This growth inhibition required the E2F-binding pocket domain but not the cyclin-binding domain of p130. Furthermore, increased amounts of the p130-controlled E2F, termed E2F-4, potentiated the mitogenic response of the cells to cytokine and the constitutive overexpression of E2F-4 rendered the cells cytokine-independent. Our results indicate the existence of a non-pRB-based cell cycle whose operation depends primarily on the interplay between p130 and E2F-4 in certain hematopoietic cells.
In mammalian cells, the decisions regulating cell proliferation are executed through a dynamic competition between positive and negative growth signals. Cells commit to divide at the cell cycle check point termed the restriction point in late G1 phase of the cell cycle by integrating growth-related information from inside and outside the cell (1). When the intracellular or extracellular environment is not satisfactory for proliferation, cells reset the cell cycle program and return to a quiescent state, called G0 phase.
The retinoblastoma tumor suppressor protein (pRB) is a nuclear phosphoprotein ubiquitously expressed in somatic cells. It acts as a potent inhibitor of cell growth and is thought to play a crucial role in the decision-making at the restriction point (2, 3). The action of pRB is currently explained by its physical interaction with cellular targets, notably E2F transcription factors. Hyperphosphorylation of pRB in late G1 phase of the cell cycle by cyclin-dependent kinases (CDKs) abolishes the ability of pRB to form complexes with E2Fs. Once freed from pRB, the E2Fs are thought to provoke a wave of transcription that is essential for G1 to S phase progression and subsequent cell division (4).
pRB receives phosphorylation at multiple serine/threonine residues by G1 cyclin–CDK complexes (2, 3). The activities of CDKs are regulated at multiple levels that include cyclin binding, regulatory phosphorylation, and association with two distinct families of CDK inhibitors (CKIs) (5). One of the CKIs, p16INK4A, selectively inhibits cyclin D–CDK4/6 kinase activity through its specific binding to the CDK4 or CDK6 (6).
Two pRB-related proteins, p107 and p130, share common structural organization, termed the “pocket domain,” with pRB (7–10). These pRB family proteins interact with DNA tumor virus oncoproteins and cellular targets such as E2Fs via the pocket domain. p107 and p130 are structurally closer to each other than to pRB, suggesting the existence of functional redundancy between the two proteins. Indeed, p107 and p130 share the unique property of forming complexes with cyclin E or cyclin A in the presence or absence of CDK2, whereas pRB does not (11–13).
Recent studies have shown that each member of the pRB family proteins binds a distinct subset of E2Fs at different stages of the cell cycle. Of the five E2Fs identified to date, pRB preferentially interacts with E2F-1, E2F-2, and E2F-3, whereas p107 and p130 specifically interact with E2F-4 and E2F-5 (14–20). Accordingly, each of pRB family appears to regulate distinct E2F species and dictate distinct target gene activation (20). p107 and p130 are also phosphorylated in a cell-cycle-dependent manner and may be substrates for cyclin D-dependent kinases (21, 22). This suggests that their function is also regulated by a manner analogous to pRB.
Despite extensive structural and functional similarities among pRB family proteins, the special importance of pRB in cell cycle regulation has been suggested by the observation that cyclin D-dependent kinase activities are essential for the G1/S transition in cells possessing functional pRB but are completely dispensable in cells lacking it (23, 24). In accord with this observation, ectopic overexpression of p16INK4A inhibits cyclin D–CDK kinase activity and represses cell proliferation only in those cells that possess functional pRB (25, 26). These observations raise the possibility that pRB is the only biologically critical substrate of cyclin D–CDK4/6 complexes.
The critical role of pRB in cell growth regulation is further supported by the observation that loss of pRB function, either through genetic mutation or dysregulated phosphorylation, is widely observed in a variety of human cancers (2, 3). However, in certain cell types, such as those of hematopoietic origin, mutation of the retinoblastoma gene (RB) is rarely associated with cellular transformation (27–29). Furthermore, inactivation of pRB does not appear to increase the oncogenic potential of hematopoietic cells, suggesting differential sensitivity to pRB in cellular transformation among various cell types (30).
Accordingly, it is still uncertain whether the pRB-based cell cycle control is operative in all cell types throughout the body, as is widely presumed, or whether this control operates only in some cell types, with others having an alternative control circuitry. The operation of the latter is suggested by the existence of the pRB-related pocket proteins and the multiple E2F species. In this work, we addressed the centrality of pRB and the related pocket protein in hematopoietic cell cycle control by conditionally overexpressing them in cytokine-dependent lymphoid cells. We report evidence for an alternative mechanism of cell cycle control that does not depend on pRB.
MATERIALS AND METHODS
Construction of Plasmids.
The TcIP promoter was made from the tetracycline (Tc)-repressible promoter (31) by introducing an additional regulatory element, the Escherichia coli lac operator, adjacent to the Tc-repressible transactivator (tTA)-responsive elements. pOPTET-BSD is an inducible cDNA expression vector having the TcIP promoter and the blasticidin-resistance gene as a drug-selection marker. pOPTET-puro was constructed from pOPTET-BSD by replacing the blasticidin-resistance gene with the puromycin-resistance gene. pRBΔS/T-P was generated from a cDNA encoding pRBΔp34HA mutant (a gift from P. Hamel) (32) by oligonucleotide-mediated mutagenesis with the use of Chameleon site-directed mutagenesis system (Stratagene) according to the manufacturer’s instructions. The positions changed to alanine in pRBΔS/T-P were Ser-224, -243, -561, -601, -605, -772, -781, -786, -787, -788, -800, -804, -814, and -819 and Thr-246, -350, -364, and -367. A cDNA encoding p130(Δ620–697) (33) was a gift from P. Whyte. A cDNA for p130(Δ846–1119) was made by introducing a synthetic oligonucleotide encoding the C-terminal 20 amino acids of human p130 (that are recognized by anti-p130 antiserum) and a termination codon into the Bst1107I site of the p130 cDNA. cDNAs encoding human pRB, pRBΔp34HA, and pRBΔS/T-P were respectively inserted into the pOPTET-BSD. Human p130 cDNA was cloned into pOPTET-puro.
Generation of Stable Transfectants.
The 6–1 cell is a double transfectant of a cytokine [either interleukin-2 (IL-2) or IL-3]-dependent mouse pro-B cell line, F7 (34), made by successively transfecting pUHD15–1, an expression vector for tTA (31), and p3’SS, an E. coli lac repressor expression vector (35). Cells were maintained in RPMI 1640 medium containing 10% fetal calf serum and 20% WEHI-3B-conditioned medium as a source of IL-3 (20%WEHI) or recombinant IL-2 (128 units/ml). For stable transfection, 1 μg of the linearized vector DNA and 20 μg of carrier DNA were transfected into 1 × 107 6–1 cells by electroporation. Transfected cells were selected 48 h after transfection by adding blasticidin S (20 μg/ml) or puromycin (1.5 μg/ml) to the culture medium containing Tc (1 μg/ml). Two to 3 weeks after transfection, drug-resistant cells were cloned as single cells by the limiting dilution method.
Protein Induction and Immunoblot Analysis.
Stable transfectants were cultured in medium supplemented with Tc (1 μg/ml) to suppress cDNA expression. For protein induction, cells were washed twice with PBS and cultured in medium containing 5 mM isopropyl β-d-thiogalactopyranoside (IPTG) without Tc for 24–48 h before cell lysates were prepared. Cells were lysed in ELB buffer (36) and cell lysates were loaded on a 7.5% SDS/polyacrylamide gel and blotted onto poly(vinylidene difluoride) (PVDF) membrane filter. Proteins were visualized with the ECL detection system (NEN). Antibodies used for immunoblot analysis were purchased from Santa Cruz Biotechnology except anti-pRB (product 14001A, PharMingen); anti-p130 (product sc-317), anti-E2F-4 (product sc-866), and anti-HA (product sc-805).
Electromobility Shift Assays.
E2F–DNA binding assay was performed as described (18) with a minor modification to improve the separation of each complex; whole cell lysates were cleared by centrifugation at 100,000 × g for 30 min at 4°C before binding reactions. Antibodies used for supershift assays were purchased from Santa Cruz Biotechnology except anti-pRB(product 14051A, PharMingen), anti-E2F-1(product sc-193x), anti-E2F-2 (product sc-633x), anti-E2F-3 (product sc-878x), anti-E2F-4 (product sc-1082x), anti-E2F-5 (product sc-1083x), anti-p107 (product sc-318), and anti-p130 (product sc-317).
IL-2 Dose–Response Analysis.
Cells were growth-arrested in G0/G1 phase by culturing with IL-2 (0.1 unit/ml) in the presence of Tc or IPTG. After 24 h, 1 × 104 cells were restimulated with various concentrations of IL-2 for 24 h and pulse-labeled with 1 μCi of [3H]thymidine (1 Ci = 37 GBq) for the last 4 h as described (34).
Cell Growth Curve.
Cells were cultured in medium containing recombinant IL-2 (128 units/ml) or 20%WEHI as an IL-3 source in the presence of Tc or IPTG. At 24-h intervals, cells were harvested, and viable cell numbers were counted by the trypan-blue dye exclusion method.
RESULTS
Conditional Expression of pRB in Cytokine-Dependent Hematopoietic Cells.
To investigate the roles of pRB in hematopoietic cell cycle control, we constructed a system that allowed us to induce the expression of pRB in a highly controlled manner in hematopoietic cells. In this system, the Tc-repressible promoter was modified by introducing an additional regulatory element, the E. coli lac operator. As a result, gene expression from the modified promoter (designated the “TcIP promoter”) was strongly repressed by Tc and was potently induced by the lactose analog IPTG in cells that coexpress tTA (31) and the lac repressor (LacI) (35). A mouse Ba/F3-derived pro-B lymphoid cell line, F7, whose growth is strictly dependent on either IL-2 or IL-3, was chosen to examine the effect of pRB on cytokine-dependent cell cycle progression (34). To make the TcIP promoter active and regulatable, expression vectors for tTA and LacI were sequentially transfected into F7 cells and a transfected clone, 6–1, that stably coexpresses tTA and LacI was established.
Effect of pRB on Cytokine-Triggered Cell Cycle Entry and Progression.
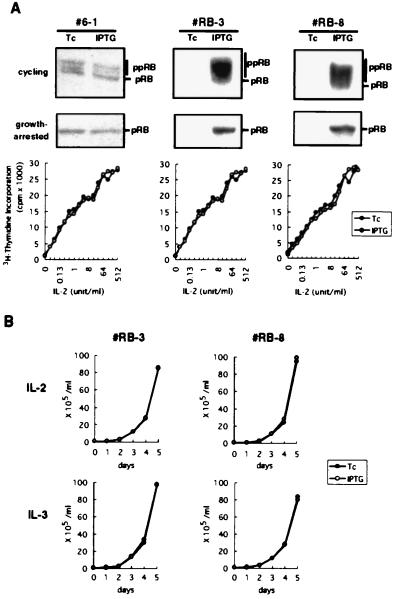
The 6–1 cells expressed endogenous pRB at levels comparable to those expressed in mouse spleen cells (data not shown). A cDNA encoding full-length human pRB was connected downstream of the inducible TcIP promoter and was transfected into 6–1 cells. After drug selection, we established more than 10 stably transfected clones that conditionally express the cDNA-encoded pRB. Of these, two clones, RB-3 and RB-8, were arbitrary chosen and subjected to detailed analysis. Levels of pRB in these pRB-transfectants cultured in the presence of Tc were almost identical to those detected in the parental 6–1 cells, suggesting that the promoter was strongly repressed by Tc. This indicates that, during the establishment of transfectants, these cells were not affected by the ectopic pRB. However, treatment of the RB-3 and RB-8 cells with IPTG after removal of Tc gave a robust induction of pRB within 24 h (Fig. 1A Top).
Figure 1.
Expression of pRB in cytokine-dependent hematopoietic cells. (A) Inducible expression of pRB in asynchronously growing (Top) or growth-arrested (Middle) 6–1 pro-B cells and two pRB-transfectant clones, RB-3 and RB-8, was examined by anti-pRB immunoblotting. The signals for endogenous pRB in 6–1 cells were obtained after prolonged exposure (10 times that of the other cells). Positions of the hypophosphorylated (pRB) and the hyperphosphorylated (ppRB) forms were indicated. (Bottom) The growth-arrested cells were then restimulated with various doses of IL-2 for 24 h and IL-2-dependent cell proliferation was evaluated by measuring [3H]thymidine incorporation into DNA. (B) The pRB-transfectants were cultured in medium containing IL-2 or IL-3 in the presence of Tc or IPTG, and cell numbers were counted.
In cells where the pRB-based cell cycle control operates, accumulation of the functional, hypophosphorylated form of pRB should block or at least retard S-phase entry until mitogenic signals succeed in neutralizing the pRB. Hence, the pRB transfectants were cultured in medium containing extremely low levels of IL-2 (0.1 unit/ml) as a survival (or anti-apoptotic) factor. Under these cytokine-starved conditions, cells arrested in the G0/G1 phase of the cell cycle within 24 h without showing any apoptotic changes (data not shown). Upon IPTG treatment, the growth-arrested cells expressed approximately 50-fold higher levels of the hypophosphorylated pRB than did uninduced cells (Fig. 1A Middle). This result demonstrates that the cDNA expression system efficiently induces pRB in cycling and growth-arrested states. The growth-arrested cells were then restimulated with various concentrations of IL-2 and DNA synthesis was monitored by measuring [3H]thymidine incorporation. As shown in Fig. 1A Bottom, the accumulated hypophosphorylated pRB failed to block or even retard the cytokine-triggered cell cycle entry and progression. Consistent with this, pRB did not significantly alter the IL-2- or IL-3-dependent cell growth despite its continued overexpression (Fig. 1B). This lack of growth inhibition by pRB might be due to the fact that virtually all of the expressed pRB was neutralized by CDK-dependent phosphorylation that was induced in response to cytokine. Alternatively, these cytokine-dependent lymphoid cells might be able to proceed through the cell cycle independent of the configuration of pRB. In either case, the result clearly demonstrated that pRB level per se is not a critical factor in determining cellular responsiveness to cytokine.
Effect of Phosphorylation-Resistant pRB on Cytokine-Dependent Cell Growth.
To address whether 6–1 cells are capable of progressing through the cell cycle in a pRB-insensitive manner, we used a phosphorylation-resistant pRB mutant, pRBΔp34HA, that was made by introducing artificial mutations into eight CDK consensus phosphorylation sites in mouse pRB (32). The mutant pRB has been shown to form stable complex with E2F-1 in cells that possess uncontrolled pRB kinase activity because of the lack of p16INK4A (37, 38).
Ectopic expression of wild-type pRB in SAOS-2 human osteosarcoma cells, which lack functional pRB, is known to induce G0/G1-phase cell cycle arrest that is associated with a large flat cell phenotype (36). This pRB-induced flat cell formation is blocked by coexpressing cyclin E or adenovirus E1A (36). Introduction of the pRBΔp34HA mutant into SAOS-2 cells also induced efficient flat cell formation. However, in contrast to the effects of wild-type pRB, the flat cell induction by the pRBΔp34HA mutant was totally resistant to cyclin E coexpression, indicating that pRBΔp34HA is highly resistant to CDK-mediated phosphorylation and associated inactivation (data not shown).
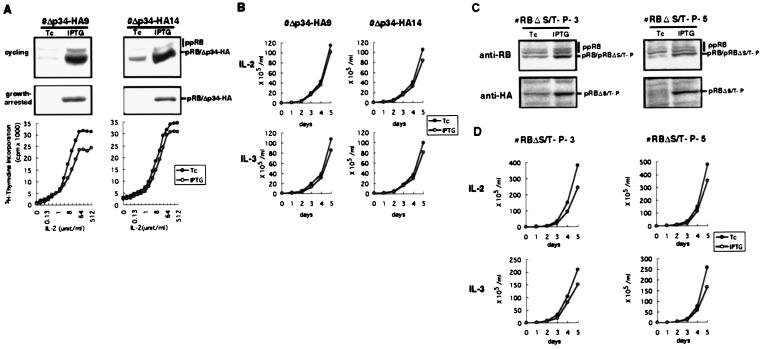
Stable transfectants that conditionally overexpress the pRBΔp34HA mutant were established from 6–1 cells. On IPTG treatment, the transfectants, Δp34-HA9 and Δp34-HA14, expressed pRBΔp34HA at approximately 50-fold higher levels than the endogenous pRB in both cycling and growth-arrested states (Fig. 2A Top and Middle). The growth-arrested transfectant cells were then restimulated with various doses of IL-2 and DNA synthesis was monitored by [3H]thymidine incorporation into DNA. Despite its phosphorylation-resistance, the PRBΔp34HA mutant had only a marginal inhibitory effect on 6–1 cell cycle entry and progression (Fig. 2A Bottom) and cells started to grow upon IL-2 or IL-3 stimulation despite the presence of pRBΔp34HA (Fig. 2B).
Figure 2.
Effect of phosphorylation-resistant pRB mutants on cytokine-dependent 6–1 cell growth. (A) Induction of pRBΔp34HA in cycling (Top) or growth-arrested (Middle) transfectants, Δp34HA-9 and Δp34HA-14, was examined by anti-pRB immunoblotting. Positions of the hypophosphorylated pRB (pRB), the phosphorylation-resistant pRB mutant (Δp34-HA), and the hyperphosphorylated pRB of endogenous origin (ppRB) were indicated. The growth-arrested cells were then restimulated with various doses of IL-2, and cell cycle entry and progression triggered by IL-2 were examined by [3H]thymidine incorporation (Bottom). (B) The pRBΔp34HA-transfectants were cultured in medium containing IL-2 or IL-3 in the presence of Tc or IPTG, and cell numbers were counted. (C) Induction of pRBΔS/T-P, which has an HA epitope at the N terminus, in asynchronously growing transfectant cells in the presence of Tc or IPTG was examined by immunoblotting with anti-pRB or anti-HA. (D) The pRBΔS/T-P-transfectants were cultured in medium containing IL-2 or IL-3 in the presence of Tc or IPTG, and cell numbers were counted.
Because the pRBΔp34HA mutant still possesses several serine and threonine residues that are a potential target of CDK, we generated another phosphorylation-resistant pRB mutant, pRBΔS/T-P, from pRBΔp34HA by using site-directed mutagenesis to replace additional 10 serine and threonine residues that constitute serine–proline and threonine–proline motifs with alanine residues. As with pRBΔp34HA, pRBΔS/T-P possesses an N-terminal hemagglutinin (HA) tag and is recognized by anti-HA antibody. Ectopic expression of pRBΔS/T-P in SAOS-2 cells also induced strong G0/G1 cell cycle arrest that was associated with large flat cell phenotype and the flat cell formation by pRBΔS/T-P was resistant to cyclin E but was sensitive to E1A (data not shown). Thus, like pRBΔp34HA, pRBΔS/T-P retained its ability to sequester E2Fs but resisted CDK-mediated inactivation.
The 6–1-derived stable transfectants, RBΔS/T-P-3 and RBΔS/T-P-5, that conditionally express the pRBΔS/T-P mutant, which possesses an HA epitope at the N terminus, were established and the inducible expression of the mutant pRB was examined by immunoblotting with anti-pRB or anti-HA (Fig. 2C). Induction of the pRBΔS/T-P mutant in the stable transfectants again gave only a weak inhibition of the cell growth, and the cells were fully capable of proceeding through the cell cycle despite continued presence of the phosphorylation-resistant pRB (Fig. 2D). We concluded from these observations that pRB has a very limited ability, if any, to regulate the growth of the hematopoietic cells, leaving open the possibility that another regulator is primarily responsible for the cell cycle control in this cell.
Effect of p130 on Cytokine-Dependent Cell Growth.
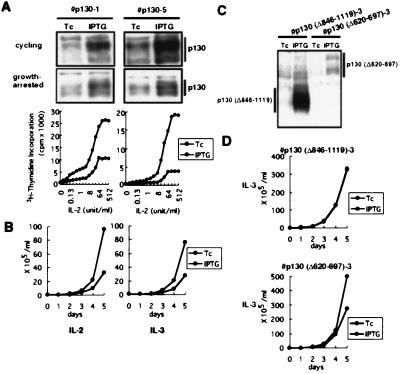
Because p130 and not p107 was predominantly expressed in the growth-arrested 6–1 cells, we next addressed the effects of the inducible ectopic expression of p130 on the growth of these cells. In striking contrast to the actions of pRB, elevated levels of wild-type p130 at 2- to 5-fold more than the endogenous levels elicited strong cell growth suppression (Fig. 3 A and B). In these p130 transfectants, cellular responses to low-doses of IL-2 (less than 8 units/ml) were also significantly impaired under culture condition where protein induction was suppressed by Tc (Fig. 3A). This was most likely due to the leaky expression of p130 from the inducible promoter despite the presence of Tc. Hence, relatively small changes of intracellular p130 levels appeared to alter cellular responsiveness to IL-2 significantly.
Figure 3.
Expression of pRB-related p130 in the cytokine-dependent 6–1 cells. (A) Induction of p130 in asynchronously growing (Top) or growth-arrested (Middle) p130-transfectants, p130–1 and p130–5, was examined by immunoblotting with anti-p130. The cells were then restimulated with various doses of IL-2 for 24 h and IL-2-dependent cell proliferation was evaluated by measuring [3H]thymidine incorporation into DNA (Bottom). (B) The p130 transfectants were cultured in medium containing IL-2 or IL-3 in the presence of Tc or IPTG, and cell numbers were counted. (C) Induction of p130(Δ620–697) mutant that lacks the cyclin–CDK binding site or p130(Δ846–1119) mutant that lacks pocket region was examined by immunoblotting with anti-p130. (D) The stable transfectants for each mutant p130 were cultured in medium containing IL-3 in the presence of Tc or IPTG, and cell numbers were counted. The same result was reproduced twice for each clone.
p130 and p107 are able to inhibit cyclin A/E-CDK2 kinase activity by direct binding (39). This raised the possibility that the p130-mediated growth inhibition was due to CDK inhibition. To test this, we conditionally expressed a p130 mutant that lacked the cyclin A/E-CDK binding region, p130(Δ620–697) (33). The mutant p130 was still capable of inhibiting the growth of the 6–1 cells (Fig. 3 C and D). In contrast, another p130 mutant, p130(Δ846–1119), which lacked the functional pocket domain but retains the CDK binding spacer region totally lost its growth-suppressing activity as examined by conditional overexpression. The result indicates that the growth inhibition was due to the pocket function of p130 and not to its ability to bind cyclin A and E.
E2F Species Expressed in the Cytokine-Dependent Cells.
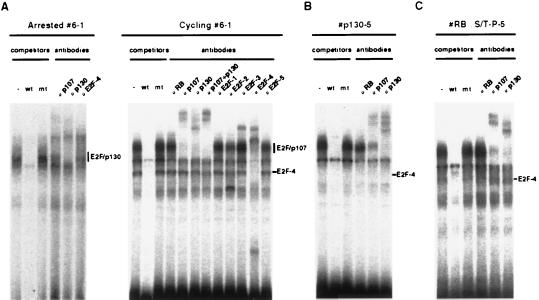
The only known proteins shown to date to specifically interact with the pocket domain of p130 in vivo are E2Fs. Of five E2F species isolated, E2F-1, E2F-2, and E2F-3 are found specifically associated with pRB. In contrast, E2F-4 and E2F-5 preferentially interact with p107 and p130 (14–17). As reported in cell types such as fibroblasts and lymphocytes (13, 18, 19), E2F-4 was predominantly expressed throughout the cell cycle in 6–1 pro-B cells and the p130/E2F-4 complex represented a prevalent form of E2F activity in the growth-arrested 6–1 cells as measured in an E2F gel shift-supershift assay (Fig. 4A). On the other hand, the activities of other E2Fs except E2F-3 were hardly detectable in both cycling and growth-arrested 6–1 cells. The p130/E2F-4 complexes detected in the growth-arrested 6–1 cells disappeared after cytokine stimulation, and free E2F-4 as well as E2F-4 complexed with p107 were readily detectable in cycling 6–1 cells (Fig. 4A). However, in the p130 transfectant that ectopically expresses p130, the induced p130/E2F-4 complex persisted despite the cytokine stimulation (Fig. 4B). This suggested that p130 inhibited the cell cycle of these cells by preventing the accumulation of free E2F-4. In contrast, in cells overexpressing pRBΔS/T-P, the phosphorylation-resistant pRB mutant could not sequester E2F-4 sufficiently enough to halt the cell cycle (Fig. 4C), potentially due to lower binding affinity or the unstable nature of the pRB/E2F-4 complex (14–17).
Figure 4.
E2F–DNA binding activity in the cytokine-dependent 6–1 cells. (A) Identification of E2F species. Whole-cell extracts prepared from growth-arrested (Left) or asynchronously growing (Right) 6–1 cells were subjected to an E2F gel shift assay. (B) Effect of p130 overexpression on E2F-DNA binding activity. Whole cell lysates were prepared from cells overexpressing p130 and were subjected to an E2F gel shift assay. Anti-p130 used in the assay cross-reacted with p107. (C) Effect of pRBΔS/T-P overexpression on E2F–DNA binding activity. Whole-cell lysate was prepared from the asynchronously growing cells overexpressing pRBΔS/T-P and were subjected to an E2F gel shift assay. Wild-type (wt) or mutant (mt) E2F gel shift oligonucleotides were used as competitors (A–C). The positions of free E2F and E2Fs complexed with pRB family proteins are indicated (A–C).
Effect of E2F-4 on Cytokine-Dependent Cell Growth.
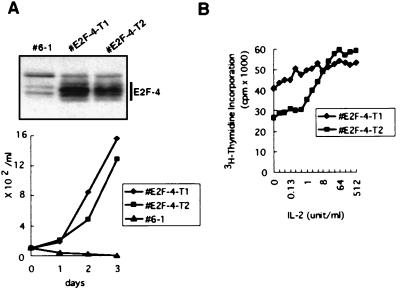
To address the role of E2F-4 in the cytokine-dependent growth of the 6–1 cells more directly, we generated 6–1-derived stable transfectants that conditionally express E2F-4 after IPTG treatment. Culturing these E2F-4 transfectants in cytokine-free medium containing IPTG allowed the cells to grow in the absence of cytokine. The cells all expressed high levels of E2F-4 and their growth was totally independent of cytokine (Fig. 5). In the control experiment, cells transfected with an empty vector never survived more than 3 days in the absence of cytokine.
Figure 5.
Effect of E2F-4 on cytokine-dependent 6–1 cell growth. (A) Stable E2F-4-transfectants, E2F-4-T1 and E2F-4-T2, were cultured in cytokine-free medium in the presence of IPTG. (Upper) Expression of ectopically introduced E2F-4 in E2F-4-T1 and E2F-4-T2 that grow in the absence of cytokine was analyzed by immunoblotting whole cell lysates with anti-E2F-4. (Lower) Cell growth curves of the E2F-4 transfectants in the absence of cytokine. As a control, parental 6–1 cell was cultured in the cytokine-free medium. (B) IL-2 dose–response experiment of the E2F-4 transfectants. E2F-4-T1 and E2F-4-T2 cells were stimulated with various doses of IL-2 for 24 h and DNA synthesis during last 4 h of the culture was evaluated by measuring [3H]thymidine incorporation.
DISCUSSION
In this work, we addressed the generality of the pRB-based cell cycle control in mammalian cells, particularly focusing on the role of pRB in cytokine-dependent proliferation of hematopoietic cells in which genetic inactivation of RB is rarely associated with the cellular transformation.
We demonstrate that in the 6–1 pro-B lymphoid cell line, the growth-promoting signals triggered by either IL-2 or IL-3 are able to initiate DNA synthesis and subsequently cell cycle progression, despite a continued presence of the functional hypophosphorylated form of pRB. We further demonstrate that this pRB-insensitive proliferation was strongly inhibited by wild-type p130. The same results were also obtained in another IL-3-dependent hematopoietic cell line, 32D, by using the same conditional expression system; IL-3-dependent growth of the 32D myeloid cells was resistant to pRB but was sensitive to p130 (data not shown), indicating that the observation is not unique to the 6–1 cells. Our work thus revealed the existence of cells whose cell cycle can progress without using pRB-controlled cellular proteins and hence argues strongly against the idea that pRB inactivation is an obligatory point of convergence for cell cycle progression in all somatic cells.
The insensitivity of the 6–1 cells to pRB indicates that critical E2F functions required for G1-cell cycle progression can be sufficiently executed by E2F species that are not under the control of pRB. Moreover, the possibility exists that 6–1 cells are insensitive to nonphosphorylatable pRB due to their lack of pRB-specific E2Fs that might be required for pRB to repress certain genes upon complex formation with pRB.
The p130-mediated growth inhibition required the pocket region of p130 that is used to interact with E2Fs but did not require the spacer region that is needed to regulate CDK2 activity. To date, the only known cellular proteins that interact with the p130 pocket region are E2F-4 and E2F-5 (14–19, 33). In 6–1 cells that had been growth-arrested by cytokine starvation or p130 overexpression, the only E2F activity detected by the gel shift assay was E2F-4 complexed with p130. This suggests that the p130-mediated cell cycle block in 6–1 cells was due to p130 binding to E2F-4 and that p130/E2F-4 complexes in the growth-arrested 6–1 cells represent a critical target that must be disrupted by cytokine signals to permit progress through the cell cycle. Accumulation of free E2F-4 or decrease of the p130/E2F-4 complex that functions as a general transcription repressor (40) may suffice to allow these cells to enter into S phase and subsequent cell cycle progression. This notion was supported by the conditional expression of E2F-4 in the 6–1 cells. Elevated levels of E2F-4 clearly potentiated the mitogenic response of the cells to cytokine. Furthermore, constitutive overexpression of E2F-4 was able to convert the cytokine-dependent cells into factor-independence. Hence, E2F-4, whose activity is negatively regulated by p130 through complex formation, is indeed a potent positive regulator of the cytokine-dependent cell cycle in the 6–1 cells. In this regard, the p130-related p107 also preferentially interacts with E2F-4 at different stages of the cell cycle. Accordingly, interactions between p107 and E2F-4 may also be involved in the regulation of the pRB-insensitive cell cycle. Although pRB does not bind E2F-4 with high affinity, E2F-4 is identified as a major binding partner of pRB in cells entering late G1 to S phases (18, 19). With this observation in mind, a slight inhibition of the 6–1 cell growth by overexpressed phosphorylation-resistant pRB may be explained by the stabilized interaction between pRB and E2F-4.
If p130 is a critical regulator of cell cycle in certain cell types, then one would expect a clear phenotype such as abnormal hematopoietic cell proliferation in mice lacking p130. However, p130 nullyzygous mice develop normally and do not manifest obvious hematological abnormalities (41). This indicates that critical functions of p130 in cell growth and differentiation may be effectively compensated by the p130-related p107 due to its largely redundant function with p130 (20). It is also possible that an as-yet-uncharacterized pocket protein recently reported (20) might function redundantly with p130.
In cell types such as fibroblasts, ectopic expression of wild-type pRB induces strong growth suppression, and inhibition of cyclin D–CDK by p16INK4A blocks G1 to S phase progression in pRB-positive cells but not in pRB-negative cells (23–26). This indicates that in such cells the p130 and p107 proteins are not critical regulators of proliferation even though they are expressed and their phosphorylation and inactivation are inhibited by ectopically expressed p16INK4A (23–26). Hence, in fibroblasts, pRB is the critical pocket protein involved in the regulation of the cell cycle progression. Our present work demonstrates that the converse situation may also be true. Thus, there are cells in which the phosphorylation of pRB is unimportant and where other pRB-related pocket proteins assume central control of proliferation.
Acknowledgments
We thank Dr. R. A. Weinberg and Dr. D. Cobrinik for valuable discussions and review of the manuscript, Dr. M. Ikeda for technical help with gel shift assay, Dr. H. Bujard for the Tc-controlled gene expression system, Dr. P. Hamel for pRBΔp34HA cDNA, Dr. P. Whyte for p130 cDNAs, Dr. C. Sardet for E2F-4 cDNA, Shionogi Pharmaceutical Co. for recombinant IL-2, and Ms. K. Tanabe for technical help. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture, Japan and by a grant from Vehicle Racing Commemorative Foundation.
ABBREVIATIONS
- pRB
retinoblastoma protein
- CDK
cyclin-dependent kinase
- CKI
CDK inhibitor
- IL
interleukin
- Tc
tetracycline
- tTA
tetracycline-repressible transactivator
- IPTG
isopropyl β-d-thiogalactopyranoside
References
- 1.Pardee A B. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 3.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 4.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 5.Harper J W, Elledge S J. Curr Opin Genet Dev. 1996;6:56–64. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 6.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 7.Ewen M E, Xing Y G, Lawrence J B, Livingston D M. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 8.Hannon G J, Demetrick D, Beach D. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Graham C, Lacy S, Duncan A M V, Whyte P. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 10.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Oncogene. 1993;8:2561–2566. [PubMed] [Google Scholar]
- 11.Ewen M E, Faha B, Harlow E, Livingston D M. Science. 1992;255:85–87. doi: 10.1126/science.1532457. [DOI] [PubMed] [Google Scholar]
- 12.Faha B, Ewen M E, Tsai L-H, Livingston D M, Harlow E. Science. 1992;255:87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- 13.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 14.Beijersbergen R L, Kerkhoven R,M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 15.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van’t Veer L J, Bernards R. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vairo G, Livingston D M, Ginsberg D. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda M, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moberg K, Starz M A, Lees J A. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurford R K, Jr, Cobrinik D, Lee M H, Dyson N. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 21.Beijersbergen R L, Carlee L, Kerkhoven R M, Bernards R. Genes Dev. 1995;9:1340–1353. doi: 10.1101/gad.9.11.1340. [DOI] [PubMed] [Google Scholar]
- 22.Mayol X, Garriga J, Grana X. Oncogene. 1995;11:801–808. [PubMed] [Google Scholar]
- 23.Tam S W, Theodoras A M, Shay J W, Draetta G F, Pagano M. Oncogene. 1994;9:2663–2674. [PubMed] [Google Scholar]
- 24.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Nature (London) 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 26.Medema R H, Herrera R E, Lam F, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preudhomme C, Vachee A, Lepelley P, Vanrumbeke M, Zandecki M, Quesnel B, Cosson A, Fenaux P. Br J Haematol. 1994;87:61–67. doi: 10.1111/j.1365-2141.1994.tb04871.x. [DOI] [PubMed] [Google Scholar]
- 28.Delmer A, Tang R, Senamaud-Beaufort C, Paterlini P, Brechot C, Zittoun R. Leukemia. 1995;9:1240–1245. [PubMed] [Google Scholar]
- 29.Jamal R, Gale R E, Thomas N S B, Wheatley K, Linch D C. Br J Haematol. 1996;94:342–351. doi: 10.1046/j.1365-2141.1996.d01-1804.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen J Z, Gorman J R, Stewart V, Williams B, Jacks T, Alt F W. Curr Biol. 1993;3:405–413. doi: 10.1016/0960-9822(93)90347-q. [DOI] [PubMed] [Google Scholar]
- 31.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamel P A, Gill R M, Phillips R A, Gallie B L. Oncogene. 1992;7:693–701. [PubMed] [Google Scholar]
- 33.Lacy S, Whyte P. Oncogene. 1997;14:2395–2406. doi: 10.1038/sj.onc.1201085. [DOI] [PubMed] [Google Scholar]
- 34.Hatakeyama M, Mori H, Doi T, Taniguchi T. Cell. 1989;59:837–845. doi: 10.1016/0092-8674(89)90607-7. [DOI] [PubMed] [Google Scholar]
- 35.Fieck A, Wyborski D L, Short J M. Nucleic Acids Res. 1992;20:1785–1791. doi: 10.1093/nar/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Z-X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Nature (London) 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 38.Hofmann F, Martelli F, Livingston D M, Wang Z. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 39.Zhu L, Harlow E, Dynlacht B D. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 40.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 41.Cobrinik D, Lee M-H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]