Abstract
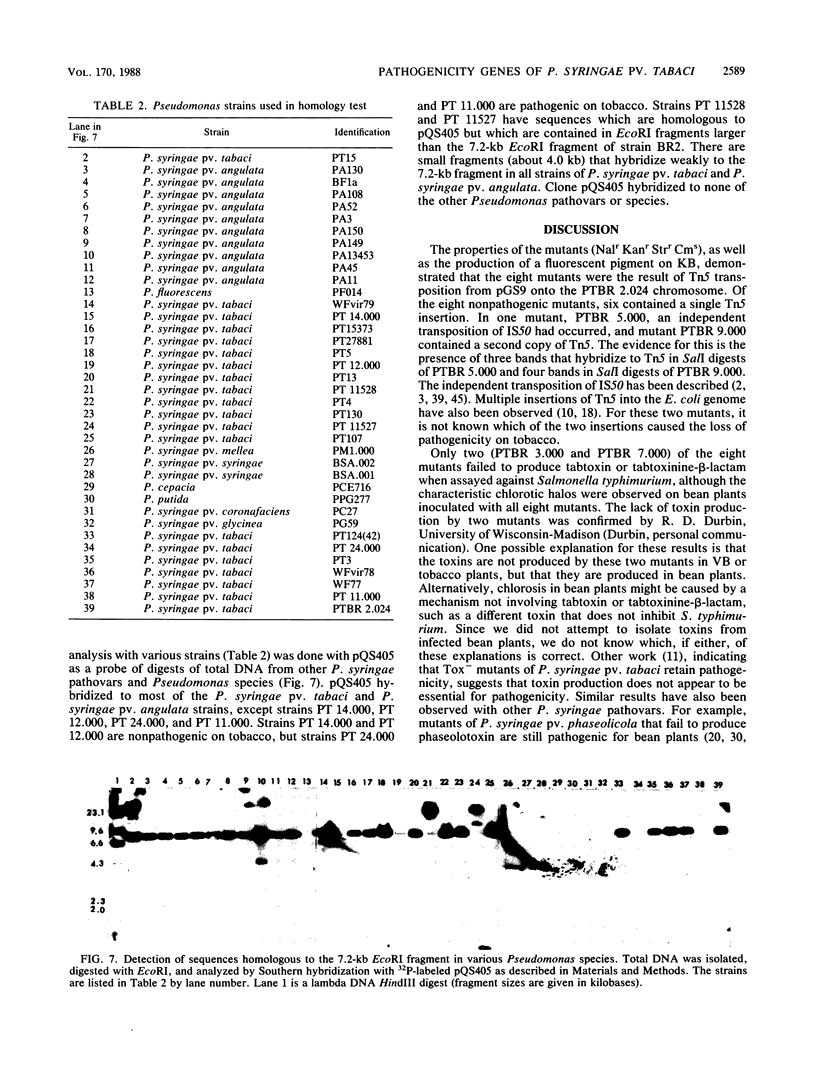
Pseudomonas syringae pv. tabaci BR2 produces tabtoxin and causes wildfire disease on tobacco and bean plants. Approximately 2,700 Tn5 insertion mutants of a plasmid-free strain, PTBR 2.024, were generated by using suicide plasmid pGS9. Of these Tn5 mutants, 8 were no longer pathogenic on tobacco plants and 10 showed reduced symptoms. All of the eight nonpathogenic mutants caused typical wildfire disease symptoms on bean plants. Two of the nonpathogenic mutants failed to produce tabtoxin. The eight nonpathogenic mutants have Tn5 insertions into different EcoRI and SalI restriction fragments. The EcoRI fragments containing Tn5 from the eight nonpathogenic mutants were cloned into vector pTZ18R or pLAFR3. A genomic library of the parent strain was constructed in the broad-host-range cosmid pLAFR3. Three different cosmid clones that hybridized to the cloned Tn5-containing fragment from one of the nonpathogenic mutants, PTBR 4.000, were isolated from the genomic library. These clones contained six contiguous EcoRI fragments (a total of 57 kilobases [kb]). A 7.2-kb EcoRI fragment common to all three restored pathogenicity to mutant PTBR 4.000. None of the six EcoRI fragments hybridized to Tn5-containing fragments from the other seven mutants. The 7.2-kb fragment was conserved in P. syringae pv. tabaci and P. syringae pv. angulata, but not in other pathovars or strains. Because the mutants retained pathogenicity on bean plants and because of the conservation of the 7.2-kb EcoRI fragment only in pathovars of tobacco, we suggest that genes on the fragment might be related to host specificity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg D. E., Johnsrud L., McDivitt L., Ramabhadran R., Hirschel B. J. Inverted repeats of Tn5 are transposable elements. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2632–2635. doi: 10.1073/pnas.79.8.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R., Vapnek D. Integration of the plasmid prophages P1 and P7 into the chromosome of Escherichia coli. J Mol Biol. 1979 May 15;130(2):161–173. doi: 10.1016/0022-2836(79)90424-8. [DOI] [PubMed] [Google Scholar]
- Daniels M. J., Barber C. E., Turner P. C., Sawczyc M. K., Byrde R. J., Fielding A. H. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 1984 Dec 20;3(13):3323–3328. doi: 10.1002/j.1460-2075.1984.tb02298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Gross D. C., Vidaver A. K. Transformation of Pseudomonas syringae with nonconjugative R plasmids. Can J Microbiol. 1981 Aug;27(8):759–765. doi: 10.1139/m81-118. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Harayama S., Palva E. T., Hazelbauer G. L. Transposon-insertion mutants of Escherichia coli K12 defective in a component common to galactose and ribose chemotaxis. Mol Gen Genet. 1979 Mar 20;171(2):193–203. doi: 10.1007/BF00270005. [DOI] [PubMed] [Google Scholar]
- Johnston J. B., Gunsalus I. C. Isolation of metabolic plasmid DNA from Pseudomonas putida. Biochem Biophys Res Commun. 1977 Mar 7;75(1):13–19. doi: 10.1016/0006-291x(77)91282-7. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lindgren P. B., Peet R. C., Panopoulos N. J. Gene cluster of Pseudomonas syringae pv. "phaseolicola" controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol. 1986 Nov;168(2):512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Obukowicz M., Shaw P. D. Construction of tn3-containing plasmids from plant-pathogenic pseudomonads and an examination of their biological properties. Appl Environ Microbiol. 1985 Feb;49(2):468–473. doi: 10.1128/aem.49.2.468-473.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet R. C., Lindgren P. B., Willis D. K., Panopoulos N. J. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. "phaseolicola". J Bacteriol. 1986 Jun;166(3):1096–1105. doi: 10.1128/jb.166.3.1096-1105.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowarski J. M., Shaw P. D. Characterization of plasmids from plant pathogenic pseudomonads. Plasmid. 1982 Jan;7(1):85–94. doi: 10.1016/0147-619x(82)90030-0. [DOI] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983 Dec;156(3):1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Transposon Tn5 specifies streptomycin resistance in Rhizobium spp. J Bacteriol. 1984 May;158(2):580–589. doi: 10.1128/jb.158.2.580-589.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt M. L., Vidaver A. K. Bacteriocin production by Pseudomonas syringae PsW-1 in plant tissue. Can J Microbiol. 1982 Jun;28(6):600–604. doi: 10.1139/m82-089. [DOI] [PubMed] [Google Scholar]
- Staskawicz B. J., Dahlbeck D., Keen N. T. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart W. W. Isolation and proof of structure of wildfire toxin. Nature. 1971 Jan 15;229(5281):174–178. doi: 10.1038/229174a0. [DOI] [PubMed] [Google Scholar]
- Taylor P. A., Schnoes H. K., Durbin R. D. Characterization of chlorosis-inducing toxins from a plant pathogenic Pseudomonas Sp. Biochim Biophys Acta. 1972 Nov 24;286(1):107–117. doi: 10.1016/0304-4165(72)90096-7. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Werneke J. M., Sligar S. G., Schuler M. A. Development of broad-host-range vectors for expression of cloned genes in Pseudomonas. Gene. 1985;38(1-3):73–84. doi: 10.1016/0378-1119(85)90205-7. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]








