Abstract
A method to identify mutations of virus proteins by using protein mass mapping is described. Comparative mass mapping was applied to a structural protein of the human rhinovirus Cys1199 → Tyr mutant and to genetically engineered mutants of tobacco mosaic virus. The information generated from this approach can rapidly identify the peptide or protein containing the mutation and, in cases when nucleic acid sequencing is required, significantly narrows the region of the genome that must be sequenced. High-resolution matrix-assisted laser desorption/ionization (MALDI) mass spectrometry and tandem mass spectrometry were used to identify amino acid substitutions. This method provides valuable information for those analyzing viral variants and, in some cases, offers a rapid and accurate alternative to nucleotide sequencing.
Identification of virus mutants currently requires sequencing part or all of the genome to determine the nature of the mutation. However, localizing the mutation prior to sequencing may be possible if the virus is well characterized biologically as well as structurally. For example, if a new disease phenotype is characterized by altered interactions of the virus with cell receptors, it is likely that a structural protein has been mutated. Thus, if the function of the viral proteins is known, a mutation can be narrowed to a particular protein or proteins.
Automated DNA sequencing is a well established method for identifying mutant proteins and is used to pinpoint specific regions that undergo mutation. In fact, high-throughput DNA sequencing often allows for expeditious identification of viral mutants and offers complete and unambiguous sequence information. However, some disadvantages of this approach include technical limitations in sequencing viral RNAs and the inability to map post-translational modifications. A complementary approach to nucleic acid sequencing of viral mutants is protein mass mapping using matrix-assisted laser desorption/ionization (MALDI) and/or electrospray ionization (ESI) mass spectrometry (1). The development of these two techniques has revolutionized the mass analysis of small and large, thermally labile biomolecules. Indeed, mass spectrometry has recently been used to characterize capsid proteins and post-translational modifications (2) and, in combination with protein digestions, has been used to investigate capsid mobility (3, 4).
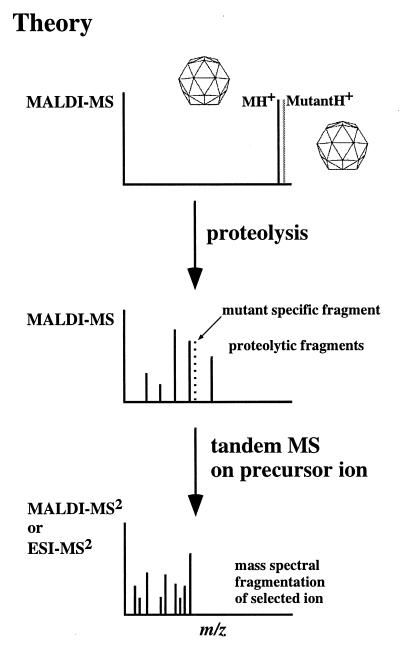
Our approach to identify mutant proteins employs protein mass mapping. Protein mass mapping consists of enzymatic digestion of a protein(s) followed by mass analysis of the resulting peptide mixture. By comparing differences in the mass of peptides that are released by such treatment, one is able to identify peptides in which amino acid differences occur. This information defines the region containing a mutation and, in cases when nucleotide sequencing is required, significantly narrows the region of the genome that must be sequenced. Accurate mass measurements and tandem mass spectrometry can then be used to definitively identify the amino acid substitution; a schematic of this approach is shown in Fig. 1.
Figure 1.
Schematic representation of the method to identify virus mutants by mass spectrometry. (Top) The observation of differences between wild-type and mutant viruses indicates amino acid changes (i.e., mutations). (Middle) Proteolysis of the viral variants generates a set of digestion fragments (one or more of which may contain the mutated amino acid), which appear at different m/z values as a consequence of an amino acid change(s). This observation provides valuable information as to the location of the mutation, augmenting its identification either by further mass spectral analysis (Bottom spectra) or nucleotide sequencing. (Bottom) Tandem mass spectrometric experiments (on the fragment containing the mutation) generate sequence information providing definitive identification of the mutated amino acid.
MATERIALS AND METHODS
Tobacco Mosaic Virus (TMV) Constructs.
The known atomic structure (5) of TMV was used to select two amino acids in the coat protein (CP) for mutation. Glu-50 and Asp-77 are located on the right slewed (RS) and right radial (RR) α-helices of the CP, respectively. The CP mutants Glu-50 → Gln, Glu-50 → Met, Asp-77 → Asn, and Asp-77 → Arg (M.B. and R.N.B., unpublished work) were generated by PCR-based site-directed mutagenesis using the plasmid pKN2 (6) containing the full-length TMV CP. The mutant CP genes were then used to replace their homologue in the TMV cDNA clone U3/12-4 (7) to generate infectious full-length cDNA clones: pTMV-Glu-50Gln, pTMV-Glu50Met, pTMV-Asp77Asn, and pTMV-Asp77Arg.
Purification of Virus Particles from Infected Plants.
Full-length transcripts were produced with T7 RNA polymerase from the wild-type pTMV (U3/12-4) and mutant pTMV cDNA clones as described previously (7) and used to inoculate 4-week-old Nicotiana tabacum Linnaeus cv. Xanthi nn plants. Ten to 15 days after inoculation, virus particles were purified from systemically infected leaves as described previously (8). Infected leaf material was ground in liquid N2 and homogenized in the extraction buffer (0.5 M Na2HPO4/0.5% sodium ascorbate). Cellular debris was removed by centrifugation and chlorophyll was removed by extraction with diatomaceous earth (grade III, Sigma). Virus particles were precipitated twice in the presence of 3% PEG8000 and 1% NaCl, washed with 5% Triton X-100, and collected by centrifugation at 90,000 × g. Virus particles were further washed (two times) with 200 mM sodium phosphate, pH 7, for 4 hr at 37°C and collected by centrifugation at 90,000 × g. The pure virus particles were then resuspended in water or in 10 mM Tris· HCl, pH 7.2/1 mM EDTA to a final concentration of 1 mg/ml and stored at 4°C. Mutant virus particles were indistinguishable from wild-type virus particles by electron microscopy (data not shown).
Production of Mutant Human Rhinovirus 14 (HRV14).
The HRV14 mutant (Cys-199 → Tyr in structural protein 1), a naturally occurring spontaneous mutant of HRV14, was selected for by isolating plaques that developed when wild-type virus was plated in the presence of 2 μg/ml WIN 52084 (9). This virus was produced from monolayers of HeLa cells infected with the Cys1199Tyr mutant. To prevent reversion, 1 μg/ml WIN 52084 was added to the medium. After incubating the infected cells for 22 hr at 37°C, virus was purified from both the cellular extract and the supernatant by using previously described protocols (10). Virus was precipitated from the supernatant by adding 7% (wt/vol) PEG8000 and 0.5 M NaCl (final concentrations) and incubating overnight at 5°C. The precipitate was harvested with 10-min centrifugation at 10,000 × g, and virus was purified from the resuspended pellets by using previously described protocols (10). Virus isolated from the cells and supernatant were combined prior to mass spectrometric analysis.
Limited Proteolysis Experiments.
Trypsin digestions of TMV variants were conducted at 25°C and trypsin digestions of HRV14 variants were conducted at 60°C, using 1 mg/ml virus in 25 mM Tris⋅HCl, pH 7.7. The enzyme-to-virus ratio (wt/wt) was adjusted to 1:100. Reaction volume was 10 μl, 0.50 μl of which was removed from the reaction, placed directly on the MALDI analysis plate, and allowed to dry before the addition of matrix (0.5 μl of 3,5-dimethoxy-4-hydroxy cinnamic acid (Aldrich) in a saturated solution of acetonitrile/water (50:50, vol/vol) with 0.25% (wt/wt) trifluoroacetic acid (TFA). MALDI–Fourier transform mass spectrometry (FTMS) experiments were conducted by using an Ionspec MALDI Fourier transform mass spectrometer, and MALDI time-of-flight (TOF) mass analyses were conducted by using a Kratos MALDI-IV and a Perceptive Biosystems Voyager Elite, both of which were equipped with delayed extraction and a nitrogen laser. External and internal mass calibration typically resulted in <5 ppm accuracy on the FTMS, allowing for the unequivocal assignment of most of the proteolytic fragments. The identity of trypsin-released fragments was determined by the Protein Analysis Worksheet (PAWS) available on the Internet.
RESULTS AND DISCUSSION
HRV14 and its naturally occurring drug-resistant mutant HRV14-Cys1199Tyr (9), together with TMV and genetically engineered mutants of TMV, were used to test the applicability of this approach to identify viral variants. Table 1 describes the HRV14 mutant, the TMV mutants, and their mass differences with respect to the capsid proteins of wild-type HRV14 and TMV strain U1.
Table 1.
HRV14 and TMV mutants and mass difference with respect to wild type
| Virus | Calculated average mass, Da | Mass difference with respect to wild-type virus, Da |
|---|---|---|
| Wild-type HRV14 | 32518.5* | — |
| HRV14-Cys199Tyr | 32601.6 | +83 |
| Wild-type TMV | 17623.7 | — |
| TMV-Glu50Gln | 17622.7 | −1 |
| TMV-Asn77Asp | 17622.7 | −1 |
| TMV-Glu50Met | 17625.7 | +2 |
| TMV-Asn77Arg | 17663.7 | +41 |
VP1 protein only.
HRV, a member of the picornavirus family, is a nonenveloped, spherical virus whose small, 280-Å diameter, shell encapsidates a plus-strand RNA genome. The viral proteins, VP1–VP3, are the major components of the icosahedral shell, whereas VP4 lies at the capsid/RNA interface (11). The 6,395-base genome of wild-type TMV-U1 is encapsidated by rod-shaped particles formed by the assembly of a single capsid protein (mass of 17,623.7 Da) in a helical structure.
Mutants.
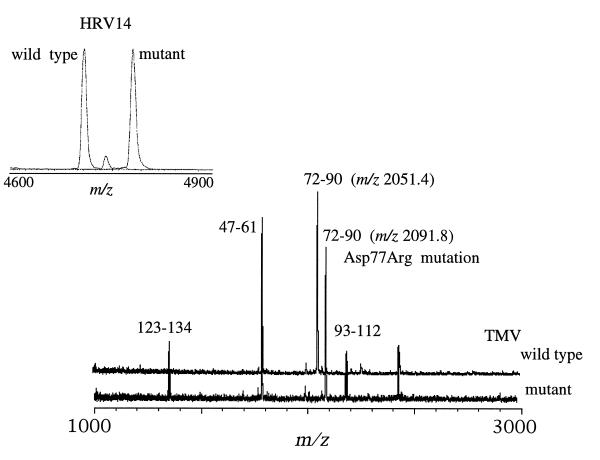
The naturally occurring mutant HRV14-Cys1199Tyr contains the Cys → Tyr amino acid change at residue 199 in the VP1 protein (9). Close comparison of MALDI mass spectra resulting from the digestion of wild type HRV14 and the HRV14-Cys1199Tyr mutant with trypsin (Fig. 2 Inset) showed most ion signals to be common to both spectra, with the exception of the ion signal at m/z 4700.5 Da (Fig. 2 Inset). This signal corresponds to residues 187–227 of the VP1 in the wild-type sequence. The corresponding ion signal in the spectrum of the mutant is observed at m/z 4783.5 Da (Fig. 2 Inset), a difference in mass of 83 Da accordant with a Cys → Tyr amino acid change (the only possible mutation with a mass difference of 83 Da). Because the sequence of this fragment contains only one Cys, at residue 199, this residue is more than likely the site of mutation.
Figure 2.
Comparison of the MALDI-TOF mass spectra resulting from the trypsin digestion of wild-type TMV and its mutant TMV-Asp50Arg. Most ion signals are common to both spectra, with the obvious exception of the signals observed at m/z 2050.5 Da in the wild-type spectrum and m/z 2091.8 Da in the mutant’s spectrum. Both signals represent residues 72–90 with the incorporation of the Asp → Arg amino acid change in the mutant. The mutation produces an ion signal (m/z 2091.8) 41 Da higher in mass than the corresponding signal from the wild-type virus. (Inset) Spectra from the trypsin digests of wild-type HRV14 and its naturally occurring mutant HRV14-Cys1199Tyr. Similarly with the HRV14 spectra, most fragments are common to both spectra, with the exception of the ion signal at m/z 4700.5 Da and 4783.8 Da in the HRV14 and HRV14-Cys1199Tyr spectra, respectively. Both ion signals corresponds to residues 187–227 of the VP1 capsid protein, and the mass difference of 83 Da between the two variants is consistent with a Cys → Tyr mutation. Mass analyses of the digestion mixtures resulted in the localization of the mutations in only a matter of minutes (<20 min, including the digestion).
Similarily, comparison of the MALDI mass spectra resulting from the trypsin digestion of wild-type TMV with that of the mutant, TMV-Asp77 → Arg (Fig. 2), shows an obvious difference in mass between the two proteins at m/z 2051.4 Da in the wild-type spectrum (Fig. 2 upper trace) and at m/z 2091.8 Da in the spectrum of the mutant (Fig. 2 lower trace). Both ion signals correspond to amino acids 72–90; however, because of the Asp → Arg mutation at residue 77, this fragment is higher (2091.8 Da) in mass than the corresponding wild-type peptide. These results were obtained in less than 20 min (including digestion time). The identification of the amino acid change requires further analyses either by tandem mass spectrometric experiments (see below) or nucleotide sequencing.
The mass spectra shown in Fig. 2 were generated by using a MALDI mass spectrometer equipped with a TOF mass analyzer and delayed extraction (12), providing mass accuracy on the order of 0.05% (i.e., ±1.0 Da on a 2000-Da peptide) and resolution of 1500 (1). Resolution is the ability of the mass spectrometer to distinguish between ions of different mass to charge ratios (m/z) and is defined by the equation r = M/ΔM, where M is the mass of the ion and ΔM is the full width of the peak at half maximum. A resolution of 1500 is easily capable of distinguishing the 41- and 83-Da mass differences on a peptide (Fig. 2); however, such a resolution is not sufficient to distinguish ion signals (on 2000-Da peptides) with a mass difference of 1 to 2 Da with a high degree of certainty. Distinguishing such small mass differences requires the use of an HPLC instrument such as a FTMS, which provides a mass resolution on the order of 20,000 or greater and an accuracy of 0.001% (13).
High-Resolution MALDI FTMS Measurements.
Introduced in 1974 by Comisarow and Marshall (13), FTMS is based on the principle of a charged particle orbiting in the presence of a very stable superconducting magnetic field. The time-dependent image current generated from the orbiting ions can then be Fourier transformed to obtain the component cyclotron frequencies of the different ions, which correspond to their m/z. A number of papers on the method and applications of FTMS can be found in the literature (13–15). FTMS offers two distinct advantages for analyzing the mass of a sample: high resolution and the ability to perform tandem mass spectrometry experiments (MS2). Furthermore, the development of the external ion source, whereby the ions are made in a separate MALDI (or electrospray) source and injected into the FTMS analyzer has made FTMS a viable analytical technique (15).
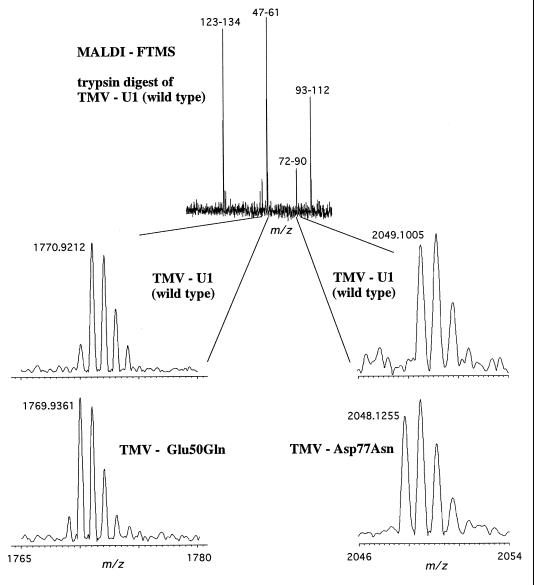
Trypsin digestion of TMV mutants TMV-Glu50Gln, TMV-Asp77Asn, and TMV-Glu50Met produced peptides that differ in mass by 1 and 2 Da compared with the wild-type viral peptides (Fig. 3). Tables 2 and 3 list the predicted and experimentally determined masses of the fragments resulting from trypsin digestion of the wild-type and the mutant TMVs. The signal observed at m/z 1770.9183 Da (Fig. 3 Upper) corresponds to a peptide that contains amino acids 47–61 with an expected m/z value of 1787.9444 Da. The difference in mass (calculated to observed) of −17 Da is due to transformation of glutamine to pyroglutamic acid (Gln → pyroGlu). This transformation occurs only when glutamine is at the N terminus of a peptide or protein (16). The corresponding fragments in the TMV-Glu50 → Gln and the TMV-GLu50 → Met (Table 3) digests were observed to have mass altered by −1 Da (Fig. 3 Left), analogous to the mutation Glu → Gln and Glu → Met, respectively. Of the 20 possible amino acids only three mutations with a mass difference of −1 Da exist, namely Glu → Gln, Asp → Asn, and Glu → Lys. The Asp → Asn mutation can be eliminated because of the absence of Asp in this fragment. The measured mass difference between the wild-type and mutant TMV-Glu50Gln was 0.9822 Da. The calculated mass difference is 0.9840 Da, whereas the calculated difference between wild type and TMV-Glu50Lys is 0.9476 Da, which is well outside the mass error observed for this instrument. Therefore, based solely on accurate mass measurement, the mutation was identified as the TMV-Glu50Gln.
Figure 3.
High-resolution MALDI FTMS mass spectra of wild-type TMV and TMV mutants. (Upper) Spectrum represents the trypsin digest of wild-type TMV-U1. The mass accuracy of these fragments was within 2 ppm with an external calibrant. (Lower) Comparison of the digests of mutants TMV-Glu50Gln (Left) and TMV-Asp77Asn (Right) with wild type reveals mass differences of 1 Da from wild type in both cases. These mass differences are accordant with the corresponding mutations (as their names indicate). The high-resolution mass measurement allowed for the localization of the mutated amino acids.
Table 2.
TMV-U1 wild-type trypsin digestion peptides (FTMS data)
| Peptide |
m/z
|
Error, ppm* | |
|---|---|---|---|
| Observed | Calculated | ||
| 47–61 | 1770.9212 | 1770.9178 | 1.9 |
| 72–90 | 2049.1005 | 2049.1020 | 0.7 |
| 93–112 | 2186.0974 | 2186.0942 | 1.5 |
| 123–134 | 1354.8083 | 1354.8059 | 1.8 |
Internal calibration.
Table 3.
Mass differences between peptides released by trypsin treatment of wild type and mutants of TMV coat proteins
| Virus | Peptide |
m/z (monoisotopic)
|
Error, ppm | Difference in mass between mutant and wild type, Da | |
|---|---|---|---|---|---|
| Observed | Theoretical | ||||
| TMV-U1 | 47–61 | 1770.9183 | 1770.9178 | 0.3* | — |
| TMV-Glu50Gln | 47–61 | 1769.9361 | 1769.9338 | 1.3* | −1 |
| TMV-Glu50Met | 47–61 | 1772.9204 | 1772.9157 | 2.7* | +2 |
| TMV-U1 | 72–90 | 2049.0962 | 2049.1020 | 2.8† | — |
| TMV-Asp77Asn | 72–90 | 2048.1255 | 2048.1180 | 3.7† | −1 |
| TMV-Asp77Arg | 72–90 | 2090.1674 | 2090.1763 | 4.3† | +41 |
Internal calibration.
External calibration.
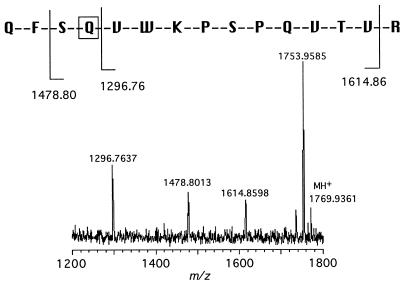
Further confirmation of the identity of the TMV-Glu50Gln mutant was obtained by tandem mass spectrometry (MS2) experiments. MS2 analysis on the trypsin fragment observed at m/z 1769.9361 Da of mutant TMV-Glu50Gln gave N-terminal and C-terminal fragmentation patterns consistent with the sequence 47–61 containing the Glu → Gln mutation (Fig. 4).
Figure 4.
Tandem mass spectral analysis of trypsin fragment 47–61 (m/z 1769.9361 Da) of mutant Glu50Gln. Both C-terminal (m/z 1641.86 Da) and N-terminal fragmentation (m/z 1296.76 and 1478.80 Da) confirm the identity of the mutation as Glu50Gln.
By comparing the mass spectrum resulting from the digestion of TMV-U1 with that of mutant TMV-Asp77Asn (Fig. 3 Lower Right) we observed a difference in mass between the ion signals corresponding to the peptides containing amino acids 72–90. This fragment was observed at m/z 2049.0952 Da in the wild-type spectrum and at m/z 2048.1255 Da in the mutant spectrum, a ΔM of −1 Da, a mass difference accordant with the Asp → Asn mutation. To confirm the identity of this mutant we considered which mutations give rise to ΔM of −1 Da. Of the three possible mutations giving a mass difference of a loss of 1 Da, Glu → Gln, Glu → Lys, or Asp → Asn, the only candidate was the Asp → Asn mutation because Glu does not occur in this fragment. In this case, however, high mass accuracy alone could not identify the mutation because fragment 72–90 contains two Asp residues. Therefore, the identity of this mutation required confirmation by either MS2 experiments or nucleotide sequencing. MS2 was attempted on the mutant; however, no data were obtained, possibly because of low concentrations of this fragment in the digestion mixture.
All trypsin digestions of the TMV mutants produced fragments containing the mutated amino acids at the outset of the reaction. However, because of tertiary/quarternary structure and other constraints on the virions, some peptide fragments are not produced as readily as others and as a result some amino acids cannot be analyzed. Such was the case in the analyses of HRV14. The fragment containing the Cys1199Tyr mutation was not readily produced when trypsin digestion of the virus was performed at room temperature. However, conducting the reactions at high temperature (60°C) readily produced the fragment of interest. Digestions performed at lower pH or in the presence of detergents can also denature the proteins and allow for more sequence coverage. Furthermore, the identity of a particular fragment or mutation can be confirmed by additional digestions with various other proteases or chemicals.
Database Searching and Viral Protein Identification.
Mass spectral data generated in this study were examined in the MS-Fit database search program (HTML page Klauser@rafael.ucsf.edu), using the NCBInr.03.21.98 database to determine how many high-accuracy mass measurements would enhance the ability to identify a virus. The TMV coat protein was easily identified by using just two tryptic fragments measured at <5-ppm accuracy (a single peptide would not allow for unequivocal identification). However, in a database search when using the average mass—i.e., the m/z values obtained on the MALDI-TOF instrument of more than 10 trypsin fragments at an accuracy of <300 ppm—the TMV capsid protein was not identified. This result demonstrates the importance of high mass accuracy when performing database searches. When the high-accuracy data from the mutants were used in the database search TMV was identified, although with a lower confidence level because the mutant fragments are not in the database. This approach could be used as a way to search the database to determine whether the virus is wild type or a previously unidentified mutant.
DNA Sequencing vs. Mass Spectrometry.
Mass analysis of the capsid proteins with or without subsequent nucleotide sequencing is an efficient approach to identifying viral variants. The purpose of this method is not to replace DNA sequencing, but to use it as a complementary tool in the characterization of viral mutants. For example, the search for antiviral agents generally leads to new drug-resistant mutants that must be characterized. In such an example the wild-type sequence is known and mass analysis may offer a rapid alternative to nucleotide sequencing, providing the site, and in some cases the identification, of the mutation. Quite often drug-resistant mutants tend to cluster at particular residues, and this technique can be used to screen for sets of unique mutants. Finally, mass analysis of the proteins provides information that cannot be obtained by nucleotide sequencing, such as the identification of post-translational modifications. Protein mass mapping also provides an alternative approach for measuring mutation rates and frequencies, in addition to being a rapid method for evolutionary studies and strain comparisons.
While the advantages of protein mass mapping to characterize viral mutants are its speed and accuracy, there are also limitations to this approach. The nanogram to microgram quantities of material and the high purity of the sample required is one limitation (detergents and buffers typically interfere with the ionization process), whereas PCR-based approaches are more sensitive and tolerant of impurities. The most serious problem is the inability of the mass spectrometer to provide complete sequence information on a protein. However, increased sequence coverage can be obtained by performing digestions under denaturing conditions. Another limitation is the inability to detect some viral structural proteins. For example, in nanoelectrospray ionization mass spectrometry experiments performed on the TMV mutants, the fragments of interest were not readily observed among the digested proteins (data not shown). In the case of MALDI, this situation may occur when especially stable viral capsids are resistant to denaturation and thus do not allow one to identify individual capsid proteins. However, the majority of viruses analyzed in this laboratory denatured under MALDI conditions, allowing for the identification of the individual capsid proteins.
Another potential problem with this approach is the identification of a mutation that does not change the mass of the peptide fragment (e.g., a Leu → Ile mutation). In addition, identification of mutants in which multiple mutations have occurred in one peptide fragment would limit the use of the procedure. The average mutation frequency of an RNA virus is 10−3 to 10−5 base substitutions per site per replication cycle (17). Thus for a 10,000-base genome, if the error frequency is 10−4, the virus will average one mutation per replication cycle. The frequency of a second mutation in the same RNA would be 10−8 base substitution per site. DNA viruses mutate at an even lower frequency because DNA polymerase generally contains proofreading capabilities. Thus, in the case of HRV14, a double mutation is highly unlikely, considering that HRV14 has a 7,500-base genome with a mutation frequency of 10−4 to 10−5 (18). Double mutations in which the mass differences cancel out could potentially be a problem when the mutations lie within the same digestion fragment. However, in cases where multiple mutations have occurred, treatment with different proteases can separate the mutated residues into different fragments, thereby allowing for their identification.
In light of these limitations, in cases where definitive mutant identification is not possible by mass spectrometric analysis alone, direct analysis of the viral proteins can still provide a wealth of information and give insight as to the region of the genome that contains the mutation. Protein mass mapping also has the ability to confirm the amino acid sequence of expressed capsid proteins. And while the focus of this study has been on the identification of a single amino acid change, this method is also well suited for detecting other forms of diversity that create new peptide fragments such as post-translational modifications and amino acid insertions/deletions.
Acknowledgments
We acknowledge Jennifer Boydston for assistance in preparation of the manuscript. This work was supported by Grants GM55775 (G.S.) and AI27161 (R.N.B.) from The National Institutes of Health, and other support was provided by the Scripps Family Chair.
ABBREVIATIONS
- MALDI
matrix-assisted laser desorption/ionization
- TMV
tobacco mosaic virus
- HRV14
human rhinovirus 14
- FTMS
Fourier transform mass spectrometry
- TOF
time-of-flight
- MS2
tandem mass spectrometry
References
- 1.Siuzdak G. Mass Spectrometry for Biotechnology. San Diego: Academic; 1996. [Google Scholar]
- 2.Siuzdak G. J Mass Spectrom. 1998;33:203–211. doi: 10.1002/(SICI)1096-9888(199803)33:3<203::AID-JMS653>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Bothner B, Dong X-F, Bibbs L, Johnson J E, Siuzdak G. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- 4.Lewis J K, Bothner B, Smith T J, Siuzdak G. Proc Natl Acad Sci USA. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namba K, Pattanyek R, Stubbs G. J Mol Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 6.Bendahmane M, Fitchen J H, Zhang G, Beachy R N. J Virol. 1997;71:7942–7950. doi: 10.1128/jvi.71.10.7942-7950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt C A, Beachy R N. Virology. 1991;181:109–117. doi: 10.1016/0042-6822(91)90475-q. [DOI] [PubMed] [Google Scholar]
- 8.Asselin A, Zaitlin M. Virology. 1978;91:173–181. doi: 10.1016/0042-6822(78)90365-3. [DOI] [PubMed] [Google Scholar]
- 9.Heinz B A, Rueckert R R, Shepard D A, Dutko F J, McKinlay M A, Fancher M, Rossmann M G, Badger J, Smith T J. J Virol. 1989;63:2476–2485. doi: 10.1128/jvi.63.6.2476-2485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson J W, Frankenberger E A, Rossmann M G, Fout G S, Medappa K C, Rueckert R R. Proc Natl Acad Sci USA. 1983;80:931–934. doi: 10.1073/pnas.80.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rueckert R R. In: Fundamental Virology. Fields B N, Knipe D M, editors. New York: Raven; 1996. pp. 609–654. [Google Scholar]
- 12.Juhasz P, Roskey M T, Smirnov I P, Haff L A, Vestal M L, Martin S A. Anal Chem. 1996;68:941–946. doi: 10.1021/ac9510503. [DOI] [PubMed] [Google Scholar]
- 13.Amster I J. J Mass Spectrom. 1996;31:1325–1337. [Google Scholar]
- 14.Comisarow M B, Marshall A G. J Mass Spectrom. 1974;31:581–585. doi: 10.1002/(SICI)1096-9888(199606)31:6<581::AID-JMS369>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, McIver R T, Jr, Hunter R L. Anal Chem. 1994;66:2077–2083. doi: 10.1021/ac00085a024. [DOI] [PubMed] [Google Scholar]
- 16.Creighton T E. Proteins. New York: Freeman; 1984. [Google Scholar]
- 17.Domingo E, Holland J H. In: The Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 161–184. [Google Scholar]
- 18.Sherry B, Mosser A G, Colonno F J, Rueckert R R. J Virol. 1986;57:246–257. doi: 10.1128/jvi.57.1.246-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]