Abstract
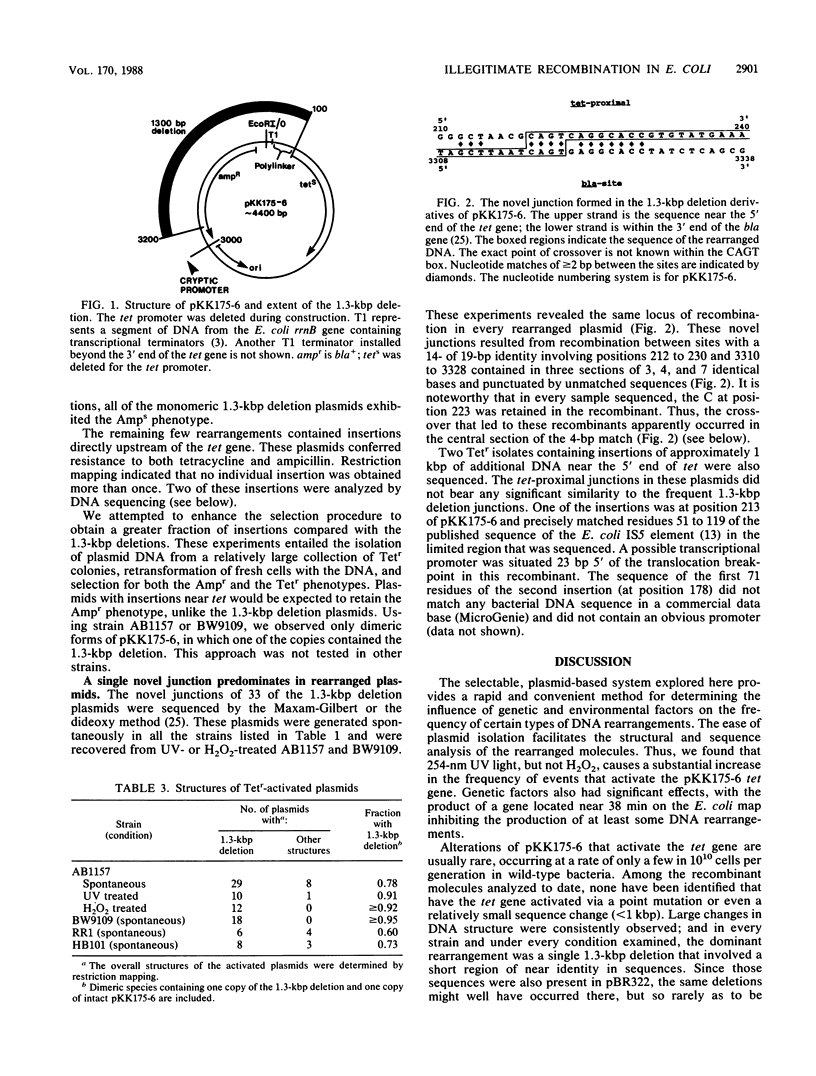
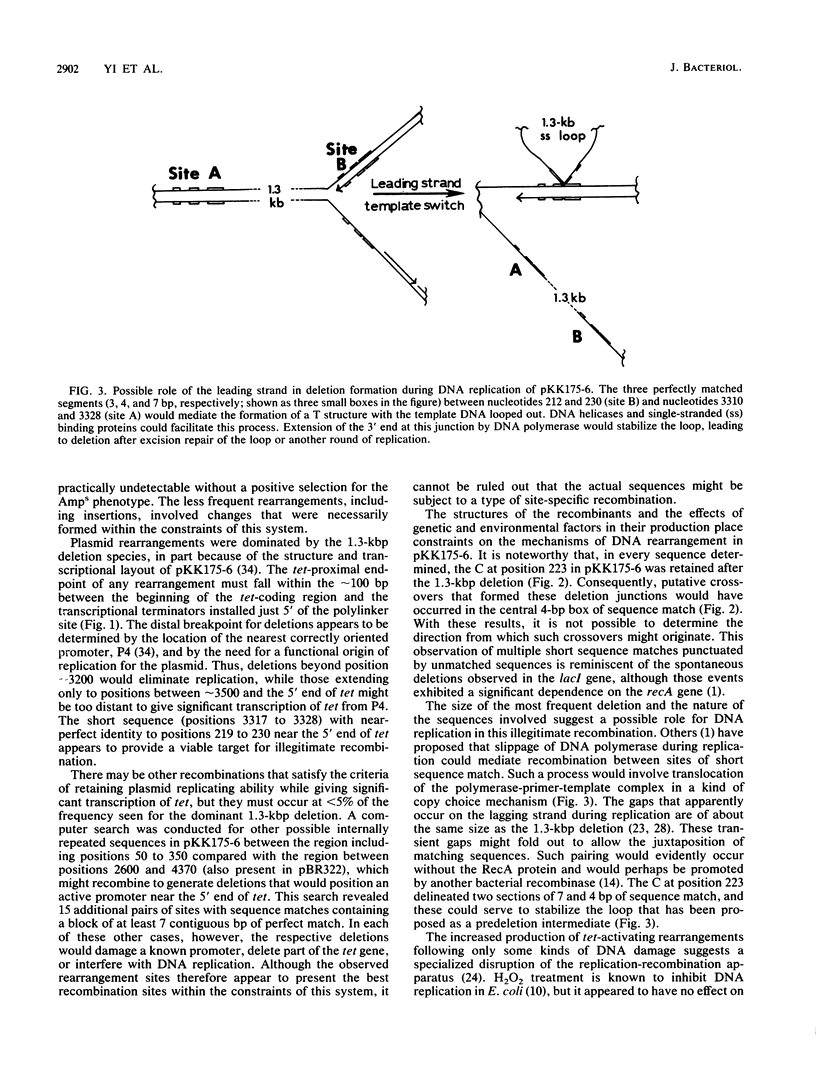
We studied DNA rearrangements in Escherichia coli by using a plasmid-based system with a transcriptionally silent tet gene and selecting for Tetr isolates. The predominant activating event was a 1.3-kilobase-pair deletion in the plasmid between two sites, with 14 of 19 base pairs being identical. These deletions occurred equally frequently in a recA+ strain and a recA13 mutant. However, the frequency of Tetr occurrence was stimulated 50-fold by treatment of the cells with UV light in a process that was at least partly independent of the SOS response. Bacterial mutants deleted for the xth-pnc region of the chromosome exhibited a strongly elevated spontaneous frequency of Tetr isolates, all with the same 1.3-kilobase-pair deletion. This phenotype of high-frequency deletion could be complemented by an episome covering this region, but not by the cloned xth gene. These studies helped to define the role of different DNA damages in illegitimate recombination and identify a region of the E. coli chromosome that contains a gene whose product normally suppresses illegitimate deletions.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem chromosomal duplications in Salmonella typhimurium: fusion of histidine genes to novel promoters. J Mol Biol. 1978 Feb 15;119(1):147–166. doi: 10.1016/0022-2836(78)90274-7. [DOI] [PubMed] [Google Scholar]
- Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984 Feb;27(2):151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Cairns J. The origin of human cancers. Nature. 1981 Jan 29;289(5796):353–357. doi: 10.1038/289353a0. [DOI] [PubMed] [Google Scholar]
- Cerutti P. A. Prooxidant states and tumor promotion. Science. 1985 Jan 25;227(4685):375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Cory S., Adams J. M. Transposition of the immunoglobulin heavy chain enhancer to the myc oncogene in a murine plasmacytoma. Cell. 1985 Jan;40(1):71–79. doi: 10.1016/0092-8674(85)90310-1. [DOI] [PubMed] [Google Scholar]
- Dean M., Park M., Vande Woude G. F. Characterization of the rearranged tpr-met oncogene breakpoint. Mol Cell Biol. 1987 Feb;7(2):921–924. doi: 10.1128/mcb.7.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Halbrook J., Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol. 1983 Feb;153(2):1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Johnson A., Fung D. Exonuclease III and endonuclease IV remove 3' blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T., Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981 Jul 16;292(5820):269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Engler J. A., van Bree M. P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981 Aug;14(3):155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Hoffman G. R. Genetic duplications in bacteria and their relevance for genetic toxicology. Mutat Res. 1985 Jun-Jul;150(1-2):107–117. doi: 10.1016/0027-5107(85)90107-1. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Aoki K., Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H. Bacteriophage T4 DNA topoisomerase mediates illegitimate recombination in vitro. Proc Natl Acad Sci U S A. 1986 Feb;83(4):922–926. doi: 10.1073/pnas.83.4.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Livneh Z., Lehman I. R. Recombinational bypass of pyrimidine dimers promoted by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1982 May;79(10):3171–3175. doi: 10.1073/pnas.79.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham B. E., Little J. W., Mount D. W. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981 Aug 25;9(16):4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggenbass M., Caro L. Intermediates of chromosomal DNA replication in Escherichia coli. J Mol Biol. 1982 Aug 5;159(2):273–301. doi: 10.1016/0022-2836(82)90496-x. [DOI] [PubMed] [Google Scholar]
- Rogers S. G., Weiss B. Cloning of the exonuclease III gene of Escherichia coli. Gene. 1980 Nov;11(3-4):187–195. doi: 10.1016/0378-1119(80)90059-1. [DOI] [PubMed] [Google Scholar]
- Sadowski P. Site-specific recombinases: changing partners and doing the twist. J Bacteriol. 1986 Feb;165(2):341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. O., Beckwith J. R. Mutagens which cause deletions in Escherichia coli. Genetics. 1969 Feb;61(2):371–376. doi: 10.1093/genetics/61.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis J. J. The chromosomal basis of human neoplasia. Science. 1983 Jul 15;221(4607):227–236. doi: 10.1126/science.6336310. [DOI] [PubMed] [Google Scholar]


