Abstract
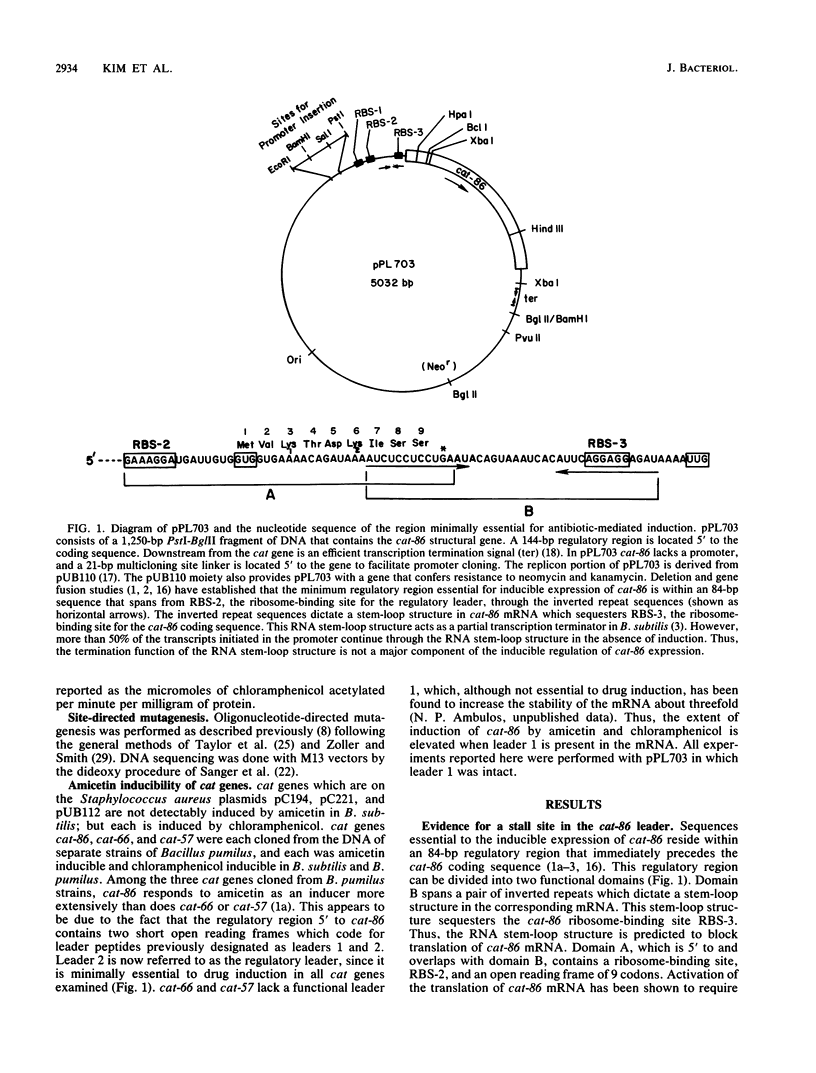
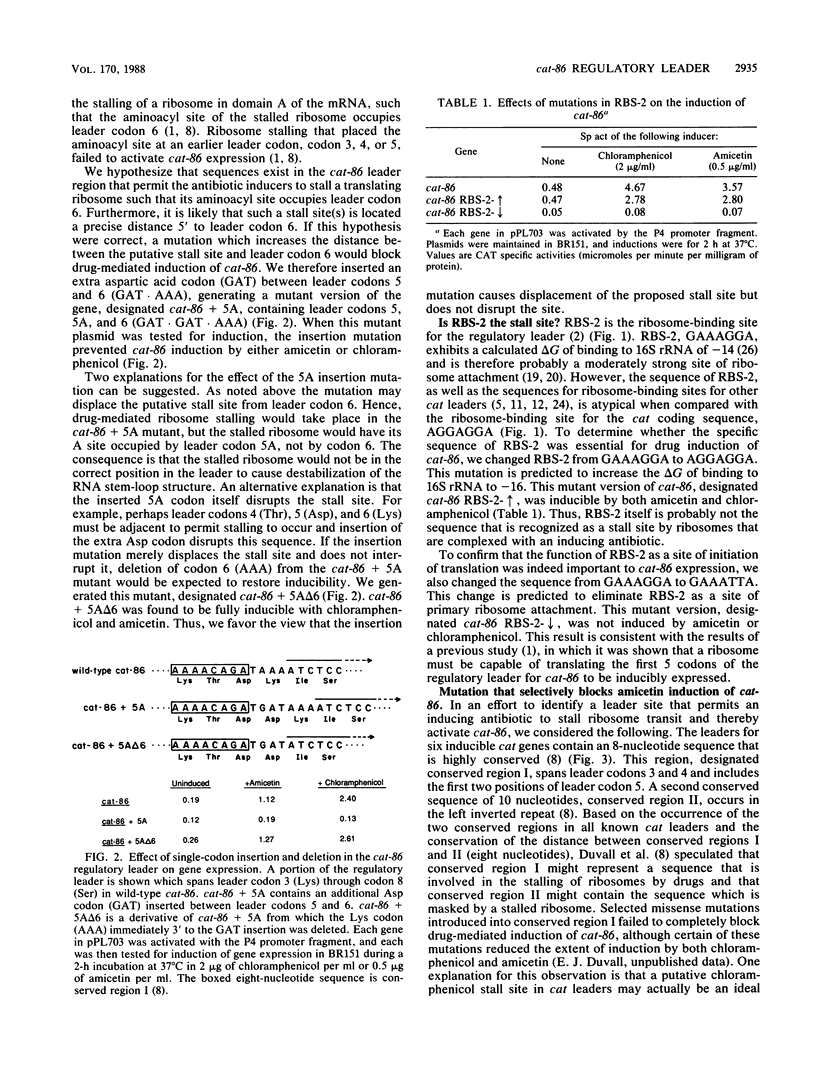
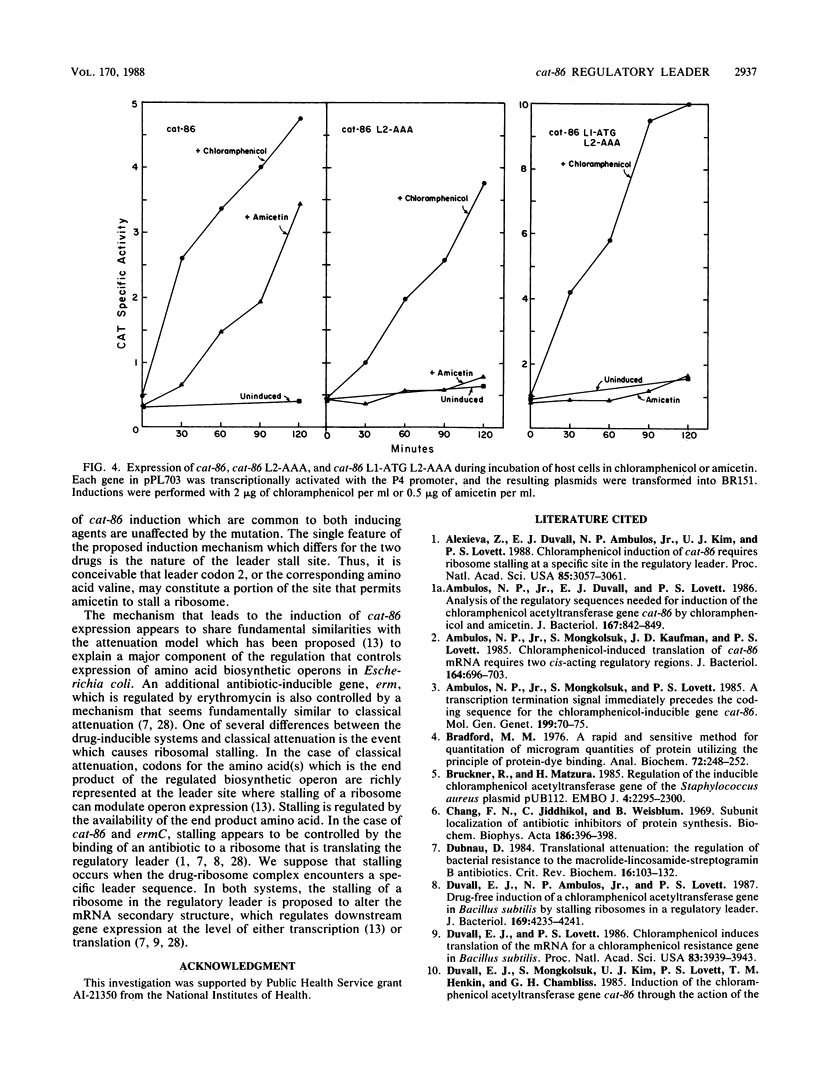
Expression of the plasmid gene cat-86 is induced in Bacillus subtilis by two antibiotics, chloramphenicol and the nucleoside antibiotic amicetin. We proposed that induction by either drug causes the destabilization of a stem-loop structure in cat-86 mRNA that sequesters the ribosome-binding site for the cat coding sequence. The destabilization event frees the ribosome-binding site, permitting the initiation of translation of cat-86 mRNA. cat-86 induction is due to the stalling of a ribosome in a leader region of cat-86 mRNA, which is located 5' to the RNA stem-loop structure. A stalled ribosome that is active in cat-86 induction has its aminoacyl site occupied by leader codon 6. To test the hypothesis that a leader site 5' to codon 6 permits a ribosome to stall in the presence of an inducing antibiotic, we inserted an extra codon between leader codons 5 and 6. This insertion blocked induction, which was then restored by the deletion of leader codon 6. Thus, induction seems to require the maintenance of a precise spatial relationship between an upstream leader site(s) and leader codon 6. Mutations in the ribosome-binding site for the cat-86 leader, RBS-2, which decreased its strength of binding to 16S rRNA, prevented induction. In contrast, mutations that significantly altered the sequence of RBS-2 but increased its strength of binding to 16S rRNA did not block induction by either chloramphenicol or amicetin. We therefore suspected that the proposed leader site that permitted drug-mediated stalling was located between RBS-2 and leader codon 6. This region of the cat-86 leader contains an eight-nucleotide sequence (conserved region I) that is largely conserved among all known cat leaders. The codon immediately 5' to conserved region I differs, however, between amicetin-inducible and amicetin-noninducible cat genes. In amicetin-inducible cat genes such as cat-86, the codon 5' to conserved region I is a valine codon, GTG. The same codon in amicetin-noninducible cat genes is a lysine codon, either AAA or AAG. When the GTG codon immediately 5' to conserved region I in cat-86 was changed to AAA, amicetin was no longer active in cat-86 induction, but chloramphenicol induction was unaffected by the mutation. The potential role of the GTG codon in amicetin induction is discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexieva Z., Duvall E. J., Ambulos N. P., Jr, Kim U. J., Lovett P. S. Chloramphenicol induction of cat-86 requires ribosome stalling at a specific site in the leader. Proc Natl Acad Sci U S A. 1988 May;85(9):3057–3061. doi: 10.1073/pnas.85.9.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Duvall E. J., Lovett P. S. Analysis of the regulatory sequences needed for induction of the chloramphenicol acetyltransferase gene cat-86 by chloramphenicol and amicetin. J Bacteriol. 1986 Sep;167(3):842–849. doi: 10.1128/jb.167.3.842-849.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Kaufman J. D., Lovett P. S. Chloramphenicol-induced translation of cat-86 mRNA requires two cis-acting regulatory regions. J Bacteriol. 1985 Nov;164(2):696–703. doi: 10.1128/jb.164.2.696-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Lovett P. S. A transcription termination signal immediately precedes the coding sequence for the chloramphenicol-inducible plasmid gene cat-86. Mol Gen Genet. 1985;199(1):70–75. doi: 10.1007/BF00327512. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brückner R., Matzura H. Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. EMBO J. 1985 Sep;4(9):2295–2300. doi: 10.1002/j.1460-2075.1985.tb03929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. N., Siddhikol C., Weisblum B. Subunit localization studies of antibiotic inhibitors of protein synthesis. Biochim Biophys Acta. 1969 Aug 20;186(2):396–398. doi: 10.1016/0005-2787(69)90020-3. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Ambulos N. P., Jr, Lovett P. S. Drug-free induction of a chloramphenicol acetyltransferase gene in Bacillus subtilis by stalling ribosomes in a regulatory leader. J Bacteriol. 1987 Sep;169(9):4235–4241. doi: 10.1128/jb.169.9.4235-4241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Lovett P. S. Chloramphenicol induces translation of the mRNA for a chloramphenicol-resistance gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3939–3943. doi: 10.1073/pnas.83.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S., Ambulos N. P., Jr, Lovett P. S. Chloramphenicol-inducible gene expression in Bacillus subtilis is independent of the chloramphenicol acetyltransferase structural gene and its promoter. J Bacteriol. 1984 Oct;160(1):1–8. doi: 10.1128/jb.160.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Duvall E. J., Lovett P. S. Transcription termination signal for the cat-86 indicator gene in a Bacillus subtilis promoter-cloning plasmid. Gene. 1985;37(1-3):83–90. doi: 10.1016/0378-1119(85)90260-4. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985 Oct 25;13(20):7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]


