Abstract
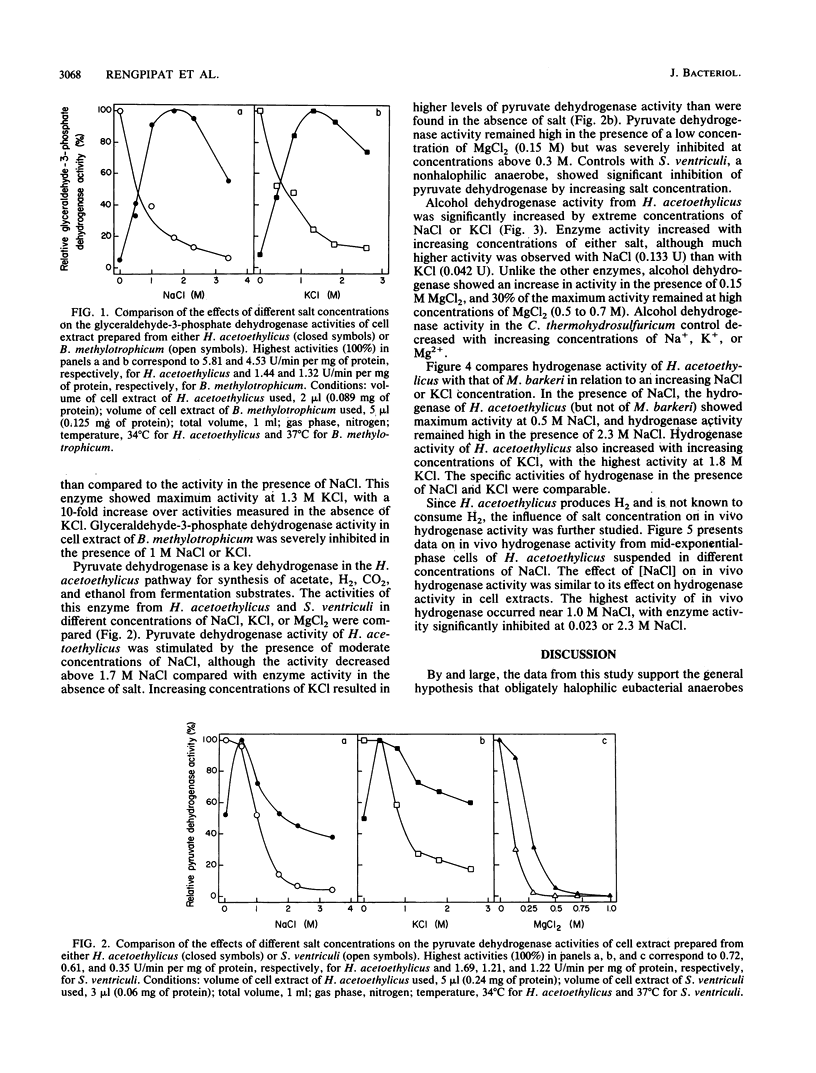
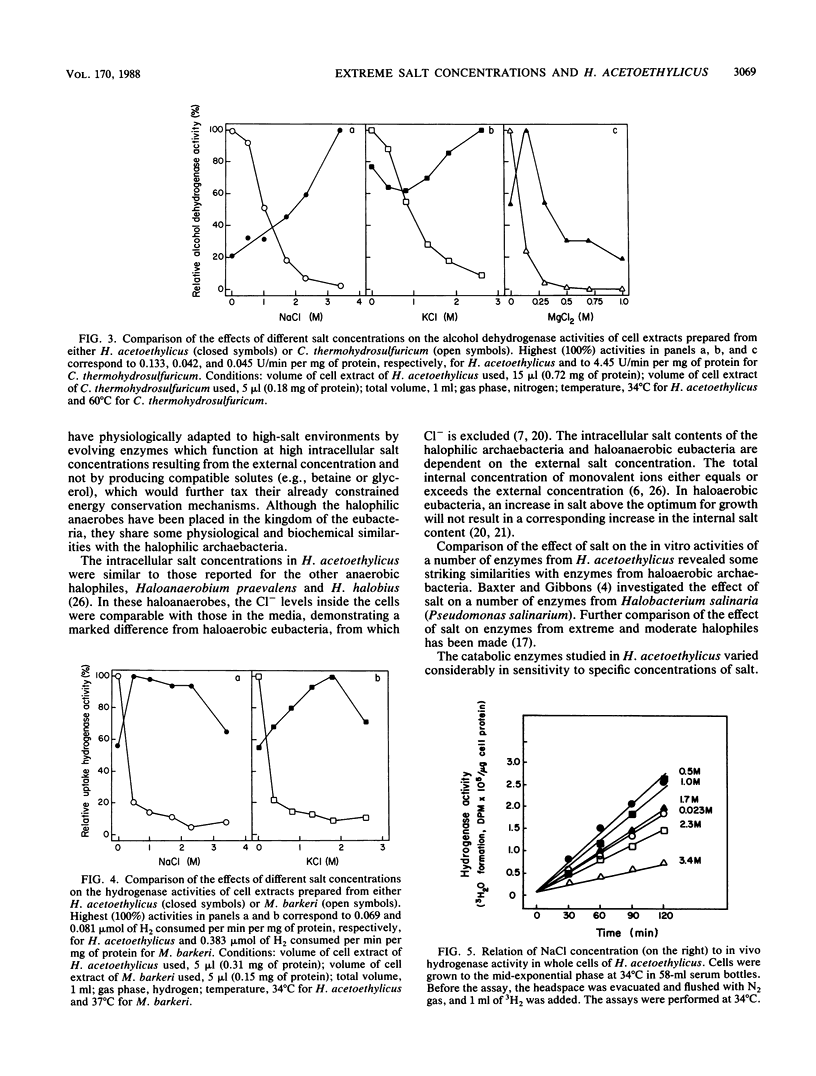
Halobacteroides acetoethylicus grew in media with 6 to 20% NaCl and displayed optimal growth at 10% NaCl. When grown in medium with an [NaCl] of 1.7 M, the internal cytoplasmic [Na+] and [Cl-] were 0.92 and 1.2 M, respectively, while K+ and Mg2+ concentrations in cells were 0.24 and 0.02 M, respectively. Intracellular [Na+] was fourfold higher than intracellular [K+]. Since Na+ and Cl- ions were not excluded from the cell, the influence of high salt concentrations on key enzyme activities was investigated in crude cell extracts. Activities greater than 60% of the maximal activity of the following key catabolic enzymes occurred at the following [NaCl] ranges: glyceraldehyde-3-phosphate dehydrogenase, 1 to 2 M; alcohol dehydrogenase (NAD linked), 2 to 4 M; pyruvate dehydrogenase, 0.5 to 1 M; and hydrogenase (methyl viologen linked), 0.5 to 3 M. These studies support the hypothesis that obligately halophilic, anaerobic eubacteria adapt to extreme salt concentrations differently than do halophilic, aerobic eubacteria, because they do not produce osmoregulants or exclude Cl-. This study also demonstrated that these halophilic, anaerobic eubacteria have a physiological similarity to archaebacterial halophiles, since Na+ and Cl- are present in high concentrations and are required for enzymatic activity.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aitken D. M., Wicken A. J., Brown A. D. Properties of a halophil nicotinamide--adenine dinucleotide phosphate-specific isocitrate dehydrogenase. Preliminary studies of the salt relations and kinetics of the crude enzyme. Biochem J. 1970 Jan;116(1):125–134. doi: 10.1042/bj1160125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAXTER R. M. An interpretation of the effects of salts on the lactic dehydrogenase of Halobacterium salinarium. Can J Microbiol. 1959 Feb;5(1):47–57. doi: 10.1139/m59-006. [DOI] [PubMed] [Google Scholar]
- BAXTER R. M., GIBBONS N. E. Effects of sodium and potassium chloride on certain enzymes of Micrococcus halodenitrificans and Pseudomonas salinaria. Can J Microbiol. 1956 Oct;2(6):599–606. doi: 10.1139/m56-072. [DOI] [PubMed] [Google Scholar]
- BAXTER R. M., GIBBONS N. E. The glycerol dehydrogenases of Pseudomonas salinaria, Vibrio costicolus, and Escherichia coli in relation to bacterial halophilism. Can J Biochem Physiol. 1954 May;32(3):206–217. [PubMed] [Google Scholar]
- Bayley S. T., Morton R. A. Recent developments in the molecular biology of extremely halophilic bacteria. CRC Crit Rev Microbiol. 1978;6(2):151–205. doi: 10.3109/10408417809090622. [DOI] [PubMed] [Google Scholar]
- CHRISTIAN J. H., WALTHO J. A. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim Biophys Acta. 1962 Dec 17;65:506–508. doi: 10.1016/0006-3002(62)90453-5. [DOI] [PubMed] [Google Scholar]
- Goodwin S., Zeikus J. G. Physiological adaptations of anaerobic bacteria to low pH: metabolic control of proton motive force in Sarcina ventriculi. J Bacteriol. 1987 May;169(5):2150–2157. doi: 10.1128/jb.169.5.2150-2157.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLMES P. K., HALVORSON H. O. PROPERTIES OF A PURIFIED HALOPHILIC MALIC DEHYDROGENASE. J Bacteriol. 1965 Aug;90:316–326. doi: 10.1128/jb.90.2.316-326.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhoff J. F., Rodriguez-Valera F. Betaine is the main compatible solute of halophilic eubacteria. J Bacteriol. 1984 Oct;160(1):478–479. doi: 10.1128/jb.160.1.478-479.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket E. R., Blanchard A. G., Metzger W. C. Proton motive force during growth of Streptococcus lactis cells. J Bacteriol. 1980 Jul;143(1):128–134. doi: 10.1128/jb.143.1.128-134.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerby R., Niemczura W., Zeikus J. G. Single-carbon catabolism in acetogens: analysis of carbon flow in Acetobacterium woodii and Butyribacterium methylotrophicum by fermentation and 13C nuclear magnetic resonance measurement. J Bacteriol. 1983 Sep;155(3):1208–1218. doi: 10.1128/jb.155.3.1208-1218.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner D. J. Halophilic bacteria. Adv Appl Microbiol. 1968;10:73–99. doi: 10.1016/s0065-2164(08)70189-8. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lanyi J. K. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev. 1974 Sep;38(3):272–290. doi: 10.1128/br.38.3.272-290.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K., Silverman M. P. The state of binding of intracellular K + in Halobacterium cutirubrum. Can J Microbiol. 1972 Jul;18(7):993–995. doi: 10.1139/m72-154. [DOI] [PubMed] [Google Scholar]
- Masui M., Wada S. Intracellular concentrations of Na+, K+, and cl minus of a moderately halophilic bacterium. Can J Microbiol. 1973 Oct;19(10):1181–1186. doi: 10.1139/m73-191. [DOI] [PubMed] [Google Scholar]
- Matheson A. T., Sprott G. D., McDonald I. J., Tessier H. Some properties of an unidentified halophile: growth characteristics, internal salt concentration, and morphology. Can J Microbiol. 1976 Jun;22(6):780–786. doi: 10.1139/m76-114. [DOI] [PubMed] [Google Scholar]
- Mullakhanbhai M. F., Larsen H. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch Microbiol. 1975 Aug 28;104(3):207–214. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- Ng T. K., Ben-Bassat A., Zeikus J. G. Ethanol Production by Thermophilic Bacteria: Fermentation of Cellulosic Substrates by Cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1981 Jun;41(6):1337–1343. doi: 10.1128/aem.41.6.1337-1343.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky T. J., Kushner D. J. Influence of temperature and salt concentration on the growth of a facultatively halophilic "Micrococcus" sp. Can J Microbiol. 1975 Jan;21(1):107–110. doi: 10.1139/m75-017. [DOI] [PubMed] [Google Scholar]
- Pugh E. L., Wassef M. K., Kates M. Inhibition of fatty acid synthetase in Halobacterium cutirubrum and Escherichia coli by high salt concentrations. Can J Biochem. 1971 Aug;49(8):953–958. doi: 10.1139/o71-138. [DOI] [PubMed] [Google Scholar]
- Schink B., Lupton F. S., Zeikus J. G. Radioassay for hydrogenase activity in viable cells and documentation of aerobic hydrogen-consuming bacteria living in extreme environments. Appl Environ Microbiol. 1983 May;45(5):1491–1500. doi: 10.1128/aem.45.5.1491-1500.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe H. D., Braselton W. E., Kaneene J. B., Slanker M. Multielement assays of bovine tissue specimens by inductively coupled argon plasma emission spectroscopy. Am J Vet Res. 1985 Mar;46(3):561–565. [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Acetate assimilation pathway of Methanosarcina barkeri. J Bacteriol. 1979 Jan;137(1):332–339. doi: 10.1128/jb.137.1.332-339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus J. G., Fuchs G., Kenealy W., Thauer R. K. Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J Bacteriol. 1977 Nov;132(2):604–613. doi: 10.1128/jb.132.2.604-613.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


