Abstract
Long chain fatty acids (LCFAs) are an important source of energy for most organisms. They also function as blood hormones, regulating key metabolic functions such as hepatic glucose production. Although LCFAs can diffuse through the hydrophobic core of the plasma membrane into cells, this nonspecific transport cannot account for the high affinity and specific transport of LCFAs exhibited by cells such as cardiac muscle, hepatocytes, and adipocytes. Transport of LCFAs across the plasma membrane is facilitated by fatty acid transport protein (FATP), a plasma membrane protein that increases LCFA uptake when expressed in cultured mammalian cells [Schaffer, J. E. & Lodish, H. F. (1994) Cell 79, 427–436]. Here, we report the identification of four novel murine FATPs, one of which is expressed exclusively in liver and another only in liver and kidney. Both genes increase fatty acid uptake when expressed in mammalian cells. All five murine FATPs have homologues in humans in addition to a sixth FATP gene. FATPs are found in such diverse organisms as Fugu rubripes, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, and Mycobacterium tuberculosis. The function of the FATP gene family is conserved throughout evolution as the C. elegans and mycobacterial FATPs facilitate LCFA uptake when overexpressed in COS cells or Escherichia coli, respectively. The identification of this evolutionary conserved fatty acid transporter family will allow us to gain a better understanding of the mechanisms whereby LCFAs traverse the lipid bilayer as well as yield insight into the control of energy homeostasis and its dysregulation in diseases such as diabetes and obesity.
Long chain fatty acids (LCFAs) are an important energy source for pro- and eukaryotes and are involved in diverse cellular processes such as membrane synthesis, intracellular signaling, protein modification, and transcriptional regulation. In developed Western countries, human dietary lipids are mainly di- and triglycerides and account for approximately 40% of caloric intake (1). These lipids are broken down into fatty acids and glycerol by pancreatic lipases in the small intestine (2); LCFAs are then transported into brush border cells where the majority is re-esterified and secreted into the lymphatic system as chylomicrons (3). Fatty acids are liberated from lipoproteins by the enzyme lipoprotein lipase which is bound to the luminal side of endothelial cells (4). “Free” fatty acids in the circulation are bound to serum albumin (5) and are rapidly incorporated by adipocytes, hepatocytes, and cardiac muscle cells. The latter derive 60–90% of their energy through the β oxidation of LCFAs (6). Although saturable and specific uptake of LCFAs has been demonstrated for intestinal cells, hepatocytes, cardiac myocytes, and adipocytes, the molecular mechanisms of LCFA transport across the plasma membrane have remained controversial (7, 8). Five proteins have been suggested to mediate LCFA uptake into cells. Four candidate LCFA transporters, FABPpm (9), 56-kDa renal FABP (10), caveolin (11), and fatty acid translocase (12), were identified by their ability to bind fatty acids. The fifth, fatty acid transport protein (FATP), was identified by Schaffer and Lodish (13) using an expression-cloning strategy. FATP is a 63-kDa plasma membrane protein which increases LCFA uptake when stably expressed in cell lines but has no effect on either glucose or short chain fatty acid transport. FATP is induced during adipocyte differentiation in vitro and is expressed in brain, skeletal muscle, heart, fat, and kidney but not liver (13). Since the liver has a large capacity for fatty acid uptake (14), we started a search for FATP homologues. Here, we describe a large family of highly homologous mammalian LCFA transporters which are widely expressed, including all tissues relevant to fatty acid metabolism. Furthermore, we also identified novel members of this family in other species including mycobacterial and nematode FATPs which, like their mammalian counterparts, are functional fatty acid transporters.
MATERIALS AND METHODS
Sequence Alignment of FATP Clones.
The DNA sequence for FATP1 was obtained from the National Center for Biotechnology Information nonredundant database. cDNAs for mmFATP2, 3, 4, and 5 were obtained by screening mouse expression libraries (purchased from GIBCO/BRL) with probes derived from the cloned expressed sequence tags (ESTs) (Research Genetics, Huntsville, AL). Full-length clones were obtained for mmFATP2 and 5 and partial sequences for FATP3 and 4. Neither FATP2 or FATP5 contains an in-frame stop codon upstream of the putative initiator methionine; initiator methionines were assigned by homology with that in mmFATP1 and by the presence of a signal sequence immediately after it. The Mycobacterium tuberculosis, Caenorhabditis elegans, and Saccharomyces cerevisiae sequences were present in the dbEST database as part of the sequencing projects for these organisms. Sequences were aligned utilizing a ClustalX algorithm and the resulting alignment exported to SeqVu. Homologous amino acid substitutions are shaded and were determined using the Dayhoff 250 method with a 50% homology cutoff.
COS Cell Transfection and LCFA Uptake.
COS cells were cotransfected using the DEAE-dextran method with the mammalian expression vector pCDNA 3.1 (Invitrogen) expressing the gene for CD2 (pCDNA-CD2) in combination with either a pCDNA 3.1 or pCMVSPORT2 (GIBCO/BRL) expression vector containing one of the murine or nematode FATP genes (pCDNA-mmFATP1, pCDNA-FATP2, pCMVSPORT-FATP5, pCDNA-ceFATPb). Two days after transfection, cells were assayed for CD2 expression with a phycoerythrin-coupled anti-CD2(PE-CD2) monoclonal antibody (PharMingen), and fatty acid uptake was assayed with a BODIPY-labeled fatty acid analogue (Molecular Probes). Briefly, cells were washed twice with PBS and stained with PE-CD2 at 4°C for 30 min in PBS containing 10% fetal calf serum. They were then washed three times with PBS/fetal calf serum for 5 min followed by an incubation for 2 min at 37°C in fatty acid uptake solution, which contained 0.1 μM BODIPY-FA and 0.1% fatty acid-free BSA in PBS (13). After 2 min, the cells were washed four times with ice-cold PBS/0.1% BSA. The cells were then removed from the plates with PBS containing 5 mM EDTA and resuspended in PBS containing 10% fetal calf serum and 10 mM EDTA. PE-CD2 and BODIPY-FA fluorescence were measured using a FACScan (Becton Dickinson). COS cells were gated on forward scatter (FSC) and side scatter. Cells exhibiting more than 300 CD2 fluorescence units representing ≈15% of all cells were deemed CD2 positive and their BODIPY-FA fluorescence was quantitated.
E. coli-Based LCFA Uptake Assay.
The full-length coding region of mtFATP and a control protein, the mammalian transcription factor TFE3, were subcloned into the inducible, prokaryotic expression vector pET (Novagen). Expression was induced with 1 mM isopropyl-β-d-thiogalactoside for 1 hr or cells were left uninduced. Cells were washed in PBS/0.1% BSA and resuspended in 1 ml PBS/0.1% BSA containing 0.1 μM [3H]palmitate (NEN) at 37°C. Uptake was stopped after the indicated incubation time by transferring the cells onto filter paper using a cell harvester (Brandel, Bethesda, MD). Filters were washed extensively with ice-cold PBS/0.1% BSA and [3H]palmitate was quantitated by scintillation counting.
Northern Blots.
Northern blot analysis of murine FATP expression was done using poly(A) mRNA blots (CLONTECH). Probes for each of the FATPs were derived from the 3′ untranslated regions of each gene and were <60% identical in sequence. Probes were labeled by random priming (Boehringer Mannheim) and hybridized at 65°C. Blots were extensively washed in 0.2% SSC/0.1% SDS at 65°C.
Generation of Phylogenetic Trees.
Complete and partial sequences for FATP genes from human, rat, mouse, puffer fish, Drosophila melanogaster, C. elegans, S. cerevisiae, and M. tuberculosis were aligned using ClustalX. A homologous region of 48 amino acids (residues 472–519 in mmFATP1) from all of the genes was used to determine phylogenetic relationships within ClustalX. Based on these data a phylogenetic tree was generated using TreeViewPPC.
RESULTS
Identification of Novel Mammalian FATPs.
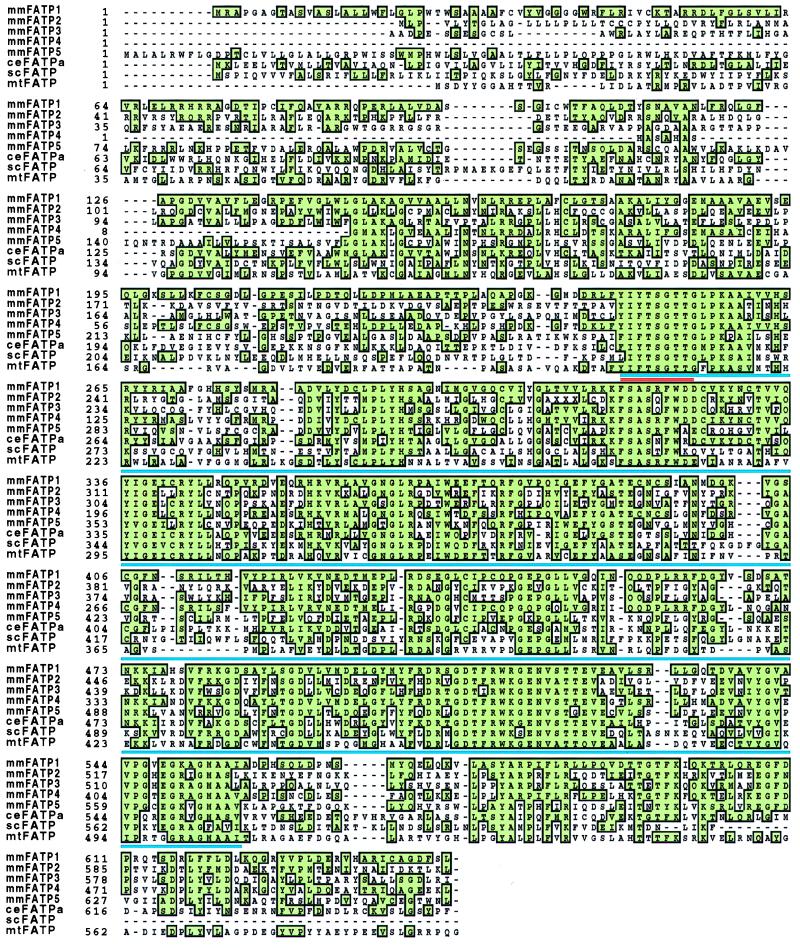
To identify novel FATPs we screened the National Center for Biotechnology Information EST database using the mouse FATP protein sequence. This strategy led to the identification of more than 50 murine EST sequences which we could assemble into five distinct contiguous DNA sequences (contigs). One contig was identical to the previously cloned FATP, which we have renamed FATP1. Another, which we have renamed FATP2, is the murine homologue of a rat gene previously identified by others as a very long chain acyl-CoA synthase (15). The other three contigs represented novel genes (FATP3, 4, and 5). By screening cDNA libraries made from mouse day 10.5 embryos and adult liver, we obtained full-length clones for FATP2 and FATP5 and nearly complete sequences for FATP3 and 4 (Fig. 1). We also identified human homologues for each of the murine genes in the EST database. Additionally, we found a sixth human gene; whether this gene is also present in the mouse will require additional studies.
Figure 1.
Sequence alignment of FATP clones.
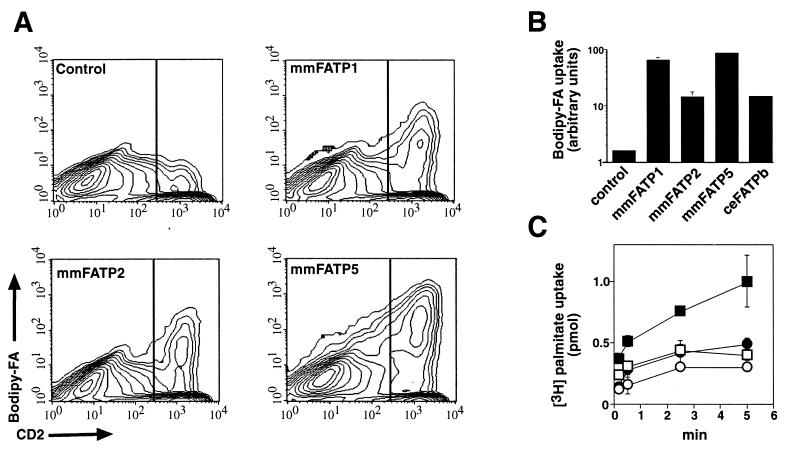
We assessed the ability of the newly identified mouse genes to function as fatty acid transporters using a fluorescence-activated cell scanning-based assay. COS cells were transiently cotransfected with expression vectors encoding the cell surface protein CD2 and either FATP1, FATP2, or FATP5, respectively. Two days after transfection, COS cells were stained with an antibody to CD2 and then incubated with a BODIPY-labeled fatty acid [BODIPY-FA, (13)]. The cells were then washed extensively, lifted off the dish, and analyzed by fluorescence-activated cell scanning. As judged by the number of CD2-positive cells, our transfection efficiency was approximately 20–30% (Fig. 2A). We quantitated fatty acid uptake in the transiently transfected COS cells by measuring the BODIPY-FA fluorescence of the CD2-positive cells. Expression of CD2 had no effect on fatty acid uptake as COS cells expressing only the transfected CD2 cDNA (CD2- positive) had the same low level of BODIPY-FA uptake as did untransfected (CD2-negative) control cells (Fig. 2A, control). In COS cells cotransfected with CD2 and FATP1, FATP2, or FATP5, uptake of BODIPY-FA by the transfected (CD2-positive) cells was increased between 15- to 90-fold over control (CD2 cDNA only) cells (Fig. 2 A and B).
Figure 2.
LCFA uptake assays. (A) COS cells were cotransfected using the DEAE-dextran method with the mammalian expression vectors pCDNA-CD2 either alone (Control) or in combination with one of the FATP-containing expression vectors (pCDNA-mmFATP1, pCDNA-mmFATP2, or pCMV-SPORT2-mmFATP5) as described in Materials and Methods. COS cells were gated on FSC and side scatter, and the results shown represent >10,000 cells. Cells exhibiting >300 CD2 fluorescence units (vertical line) representing ≈15% of all cells were deemed CD2 positive. (B) As in A, COS cells were cotransfected with pCDNA-CD2 either alone (Control) or in combination with one of the FATP-containing expression vectors (pCDNA-mmFATP1, pCDNA-mmFATP2, pCMV-SPORT2-mmFATP5, or pCDNA-ceFATPb). The mean BODIPY-FA fluorescence of the CD2-positive cells is plotted; results shown represent the average of three experiments, each consisting of >50,000 COS cells. Note that a logarithmic scale is used on the ordinate. (C) The full-length coding region of mtFATP (squares) and a control protein (TFE3; circles) were subcloned into the inducible, prokaryotic expression vector pET (Novagen). Expression was induced (solid symbols) with 1 mM isopropyl β-d-thiogalactoside for 1 hr or cells were left uninduced (open symbols). Data points were done in triplicate and counts were normalized to the number of bacteria as determined by OD600.
Expression Patterns of Murine FATPs.
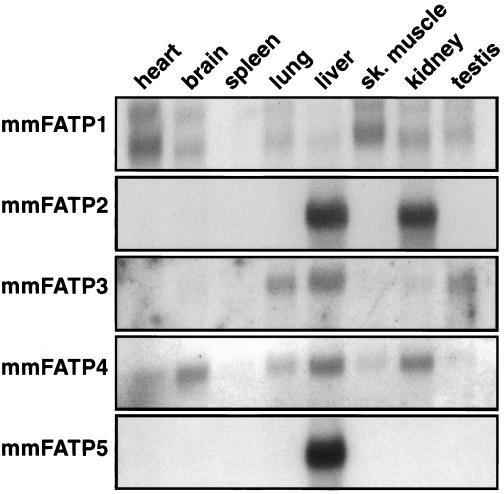
We characterized expression patterns of the murine FATP gene family by Northern blot analysis (Fig. 3); to avoid cross-hybridization, we utilized probes from the 3′ untranslated region of these genes which shared no appreciable homologies with each other. The expression pattern of FATP1 agrees with that previously found (13). FATP2 is expressed exclusively in liver and kidney, which corresponds to the reported tissue distribution of the rat homologue [very long chain acyl-CoA (VLACS)] as assessed by Western blotting (15). FATP3 is present in lung, liver, and testis; FATP4 is expressed in heart, brain, lung, liver, and kidney. FATP5 is expressed only in liver and cannot be detected in other tissues even when the blot is overexposed. The human homologue of FATP5 is also liver specific and is not expressed in a wide array of other tissues tested including fetal liver (data not shown).
Figure 3.
Northern blot analysis of murine FATP expression was analyzed using poly(A) mRNA blots (CLONTECH). Probes for each of the FATPs were derived from the 3′ untranslated repeat regions of each gene and were <60% identical in sequence.
FATPs Are Evolutionarily Conserved.
Using sequences conserved among the five murine FATP genes, we searched the EST database for FATP genes in other organisms and found two homologues in C. elegans and one in M. tuberculosis. We cloned one of the C. elegans genes from a cDNA library and expressed it in COS cells as described for the murine FATPs. Interestingly, overexpression of the nematode FATP resulted in a 15-fold increase of BODIPY-FA uptake compared with control cells (Fig. 2B). We then isolated the mycobacterial FATP gene from a phage library and assessed its ability to facilitate fatty acid uptake. E. coli transformed with a prokaryotic, isopropyl β-d-thiogalactoside-inducible expression vector containing the mycobacterial FATP gene demonstrated a significant increase in the rate of [3H]palmitate uptake after induction compared with uninduced bacteria or E. coli transformed with a control protein (Fig. 2C). Novel FATP genes were also identified in F. rubripes (puffer fish) and D. melanogaster.
Nomenclature.
We propose that the FATP genes be given a species specific prefix (mm, Mus musculus; hs, Homo sapiens; mt, M. tuberculosis; dm, D. melanogaster; ce, C. elegans, sc, S. cerevisiae) and numbered such that mammalian homologues in different species share the same number but differ in their prefix. Since the two C. elegans genes cannot be paired with a specific human or mouse FATP, we propose that they be labeled ceFATPa and ceFATPb.
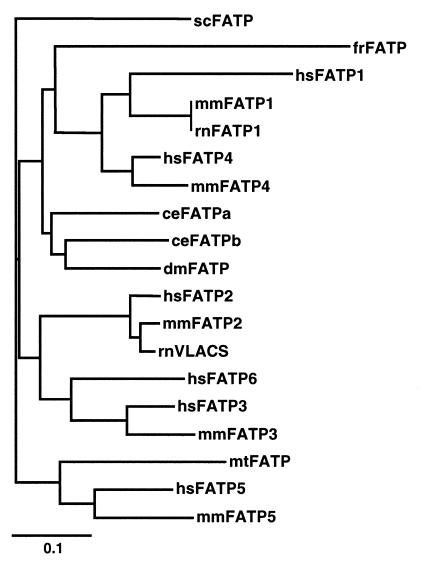
Phylogenetic Tree of FATPs.
Faergeman et al. (16) identified three regions of very strong conservation between the scFATP and mmFATP1 genes. We compared the sequences of the FATPs over a 311-amino acid FATP “signature sequence” which includes these conserved regions corresponding to amino acids 246–557 in mmFATP1 (underlined blue in Fig. 1). When compared with the National Center for Biotechnology Information nonredundant database, only one region of the “FATP signature sequence” shows significant homology to other proteins. This small stretch of amino acids (underlined red in Fig. 1) is an AMP-binding motif found in a multitude of other proteins such as acyl-CoA synthase, several CoA ligases, and gramicidin S synthetase component II (13). The relevance of this motif to fatty acid transport is unclear. Other highly conserved regions among the FATPs, including long stretches of amino acids >90% identical from mycobacteria to humans, are not found in any other class of proteins. We utilized a 48-amino acid segment of the FATP signature sequence to construct a phylogenetic tree (Fig. 4). Each of the human and mouse genes form their own branch, hsFATP6, which as yet has no murine homologue, is most closely related to hsFATP3 and mmFATP3. As expected, rnVLACS is closer in sequence to mmFATP2 than to hsFATP2. The FATP genes of invertebrates, i.e., C. elegans and D. melanogaster, are most closely related to each other. Surprisingly, the mycobacterial gene is more closely related to the human and mouse FATP5 genes than to the FATPs of any of the lower organisms. Whether this reflects coevolution of the mycobacterial and human genes awaits further study.
Figure 4.
Complete and partial sequences for FATP genes from human, rat, mouse, puffer fish, D. melanogaster, C. elegans, S. cerevisiae, and M. tuberculosis were aligned using ClustalX. Based on these data, a phylogenetic tree was generated using TreeViewPPC. The bar indicates the number of substitutions per residue, i.e., 0.1 corresponds to a distance of 10 substitutions per 100 residues.
DISCUSSION
Here, we showed that FATPs are a large evolutionarily conserved family of proteins that mediate the transport of LCFAs into cells. We identified six human and five mouse FATP genes which are expressed in a variety of tissues. These include the liver-specific gene FATP5 as well as FATP2, which is highly expressed in liver and kidney. We demonstrated that both of these proteins, as well as FATPs from nematodes and mycobacteria, are functional LCFA transporters. Interestingly, mmFATP2 is the murine homologue of a rat gene previously identified by others as a VLACS (15) even though it shares no sequence homology with other cloned acyl-CoA synthases. The rat homologue of mmFATP2 increased very long chain acyl-CoA synthase activity when expressed in COS cells (15) but this observation can be explained by our finding that expression of mmFATP2 increases the rate of fatty acid uptake from the medium; this, in turn, could stimulate transcription of a VLACS gene. Indeed, exogenously added LCFAs directly activate transcription of the long chain acyl-CoA synthase gene (17).
Not every organism utilizes a FATP to transport free fatty acids. The E. coli genome does not contain a FATP homologue; rather, the E. coli fadL gene product mediates uptake of fatty acids from the medium (18). This protein has no sequence similarity to FATPs and we cannot find homologues of fadL in the genomes of mycobacteria, yeast, C. elegans, mice, or humans.
The discovery of a diverse but highly homologous family of FATPs is reminiscent of the glucose transporter family. In a manner similar to the FATPs, the glucose transporters have very divergent patterns of tissue expression (19). The FATPs, like glucose transporters, may also differ in their substrate specificities, uptake kinetics, and hormonal regulation (20). Indeed, the levels of fatty acids in the blood, like those of glucose, can be regulated by insulin and are dysregulated in diseases such as noninsulin-dependent diabetes and obesity (21). The underlying mechanisms for the regulation of free fatty acid concentrations in the blood are not understood but could be explained by hormonal modulation of FATPs.
Another finding with potentially broad implications is the identification of a FATP homologue in M. tuberculosis. Tuberculosis causes more deaths worldwide than any other infectious agent and drug-resistant tuberculosis is re-emerging as a problem in industrialized nations (22). The de novo synthesis of fatty acids in Mycobacterium leprae is insufficient to maintain growth (23) and deletion of the scFATP gene slows their growth under certain conditions. Thus, it may be worthwhile to investigate inhibitors of mtFATP as potential therapies for tuberculosis.
Acknowledgments
We thank Drs. Monty Krieger for reading the manuscript, Jean Schaffer for the mmFATP1 cDNA, Peter Murray for the M. tuberculum library, Craig Ceol for the C. elegans library, and Fran Lewitter for expert help with the sequence analysis. This work was supported by a Program of Excellence in Molecular Biology grant from the National Heart Lung and Blood Institute (HL41484) and National Institutes of Health Grant DK 47618 (to H.F.L.). A.S. was supported by the Badische Anilinund Soda-Fabrik AG fellowship from the Studienstifung des Deutschen Volkes and D.H. by National Institutes of Health Grant 5 T32 CA 09541.
ABBREVIATIONS
- LCFA
long chain fatty acid
- FATP
fatty acid transport protein
- VLACS
very long chain acyl-CoA synthase
- EST
expressed sequence tag
- FSC
forward scatter
Footnotes
References
- 1.Weisburger J H. J Am Diet Assoc. 1997;97:S16–S23. doi: 10.1016/s0002-8223(97)00725-6. [DOI] [PubMed] [Google Scholar]
- 2.Chapus C, Rovery M, Sarda L, Verger R. Biochimie. 1988;70:1223–1234. doi: 10.1016/0300-9084(88)90188-5. [DOI] [PubMed] [Google Scholar]
- 3.Green P H, Riley J W. Aust N Z J Med. 1981;11:84–90. doi: 10.1111/j.1445-5994.1981.tb03746.x. [DOI] [PubMed] [Google Scholar]
- 4.Scow R O, Blanchette-Mackie E J. Mol Cell Biochem. 1992;116:181–191. doi: 10.1007/BF01270586. [DOI] [PubMed] [Google Scholar]
- 5.Spector A A. Clin Physiol Biochem. 1984;2:123–134. [PubMed] [Google Scholar]
- 6.Neely J R, Rovetto M J, Oram J F. Prog Cardiovasc Dis. 1972;15:289–329. doi: 10.1016/0033-0620(72)90029-1. [DOI] [PubMed] [Google Scholar]
- 7.Hui T Y, Bernlohr D A. Front Biosci. 1997;15:d222–d231. doi: 10.2741/a185. [DOI] [PubMed] [Google Scholar]
- 8.Schaffer J E, Lodish H F. Trends Cardiovasc Med. 1995;5:218–224. doi: 10.1016/1050-1738(95)00102-6. [DOI] [PubMed] [Google Scholar]
- 9.Stremmel W, Strohmeyer G, Borchard F, Kochwa S, Berk P D. Proc Natl Acad Sci USA. 1985;82:4–8. doi: 10.1073/pnas.82.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii S, Kawaguchi H, Yasuda H. J Biochem (Tokyo) 1987;101:679–684. doi: 10.1093/jb/101.3.679. [DOI] [PubMed] [Google Scholar]
- 11.Trigatti B L, Mangroo D, Gerber G E. J Biol Chem. 1991;266:22621–22625. [PubMed] [Google Scholar]
- 12.Abumrad N A, el-Maghrabi M R, Amri E Z, Lopez E, Grimaldi P A. J Biol Chem. 1993;268:17665–17668. [PubMed] [Google Scholar]
- 13.Schaffer J E, Lodish H F. Cell. 1994;79:427–436. doi: 10.1016/0092-8674(94)90252-6. [DOI] [PubMed] [Google Scholar]
- 14.Stremmel W. J Hepatol. 1989;9:374–382. doi: 10.1016/0168-8278(89)90148-7. [DOI] [PubMed] [Google Scholar]
- 15.Uchiyama A, Aoyama T, Kamijo K, Uchida Y, Kondo N, Orii T, Hashimoto T. J Biol Chem. 1996;271:30360–30365. doi: 10.1074/jbc.271.48.30360. [DOI] [PubMed] [Google Scholar]
- 16.Faergeman N J, DiRusso C C, Elberger A, Knudsen J, Black P N. J Biol Chem. 1997;272:8531–8538. doi: 10.1074/jbc.272.13.8531. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T. J Biol Chem. 1990;265:8681–8685. [PubMed] [Google Scholar]
- 18.Black P N. J Bacteriol. 1991;173:435–442. doi: 10.1128/jb.173.2.435-442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGowan K M, Long S D, Pekala P H. Pharmacol Ther. 1995;66:465–505. doi: 10.1016/0163-7258(95)00007-4. [DOI] [PubMed] [Google Scholar]
- 20.Thorens B. Am J Physiol. 1996;270:G541–G553. doi: 10.1152/ajpgi.1996.270.4.G541. [DOI] [PubMed] [Google Scholar]
- 21.Boden G. Diabetes. 1997;46:3–10. [PubMed] [Google Scholar]
- 22.Bloom B R, Small P M. N Engl J Med. 1998;338:677–678. doi: 10.1056/NEJM199803053381008. [DOI] [PubMed] [Google Scholar]
- 23.Wheeler P R, Bulmer K, Ratledge C. J Gen Microbiol. 1990;136:211–217. doi: 10.1099/00221287-136-1-211. [DOI] [PubMed] [Google Scholar]