Abstract
Peroxisomal matrix protein import requires the action of two AAA ATPases, PEX1 and PEX6. Mutations in either the PEX1 or PEX6 gene are the most common cause of the lethal neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease and account for disease in 80% of all such patients. We report here that overexpression of PEX6 can suppress the phenotypes of certain PEX1-deficient cells, that overexpression of PEX1 can suppress the phenotypes of certain PEX6-deficient cells, and that these instances of suppression are allele-specific and require partial activity of the mutated gene. In addition to genetic evidence for interaction between PEX1 and PEX6, we find that the PEX1 and PEX6 proteins interact in the yeast two-hybrid assay and physically associate with one another in vitro. We previously identified a missense mutation in PEX1, G843D, which attenuates PEX1 function and is the most common cause of these diseases, present in one-third of all such patients. The G843D mutation attenuates the interaction between PEX1 and PEX6 in both the two-hybrid system and in vitro and appears to be suppressed by overexpression of PEX6. We conclude that PEX1 and PEX6 form a complex of central importance to peroxisome biogenesis and that mutations affecting this complex constitute the most common cause of the Zellweger syndrome spectrum of diseases.
The peroxisome biogenesis disorders (PBD) are a group of lethal inherited diseases that are characterized by defects in peroxisomal protein import. Peroxisomal proteins are synthesized on free polysomes, are imported into peroxisomes post-translationally (1), and contain cis-acting peroxisome targeting signals (PTSs) that direct them into the organelle (2). To date, two PTSs have been identified: the C-terminal PTS1 (3) and the N-terminal PTS2 (4). Two distinct clinical spectra of the PBD have been recognized (5), and the most common is referred to as the Zellweger syndrome (ZS) spectrum. ZS is the most severe form of the PBD and is characterized by loss of multiple peroxisomal enzymatic functions, severe neurologic, hepatic and renal abnormalities, mental retardation, and death in early infancy. Neonatal adrenoleukodystrophy (NALD) and infantile Refsum disease represent progressively less severe forms of what is essentially the same disease. Cells from patients with any of these diseases display a defect in the import of PTS1 matrix proteins although most of these patients also display defects in PTS2 protein import (6, 7). The other clinical spectrum of the PBD is represented by classical rhizomelic chondrodysplasia punctata. This disease also is characterized by multisystem defects, mental retardation, and death in early infancy although classical rhizomelic chondrodysplasia punctata patients are defective in only PTS2 protein import (6, 7). Cell fusion complementation studies have established nine complementation groups (CG) for the Zellweger spectrum of diseases whereas classical rhizomelic chondrodysplasia punctata cells are confined to a single complementation group (8).
Extensive studies in yeast have identified 17 PEX genes whose products, peroxins, are required for peroxisome biogenesis. Of existing PEX genes, the products of all but PEX11 are required for normal peroxisomal matrix protein import (9). These include receptors for the PTS1 and PTS2 (PEX5 and PEX7, respectively) and a variety of other proteins involved in receptor docking and stability, protein translocation, and other aspects of peroxisome biogenesis. Among these are PEX1 and PEX6, two AAA ATPases required for normal import of PTS1 and PTS2 proteins and stability of PEX5 (10–12). The basic aspects of peroxisomal protein import appear to be conserved between yeast and human cells (13), and the range of phenotypes of yeast pex mutants closely mirror those of cells from patients with PBD (6, 7). Consistent with the conserved nature of peroxisome biogenesis, mutations in human homologues of yeast PEX genes have been found to cause the PBD (14, 15). Previous reports have established that PEX6 is the gene deficient in patients belonging to CG4 of the PBD (10, 16) and that mutations in PEX1 are responsible for disease in patients with PBD group 1 (12, 17). These two genes are mutated in 80% of all patients in the Zellweger spectrum, with defects in CG1 alone accounting for 65% of cases of ZS, NALD, and infantile Refsum disease (18).
During our initial identification and characterization of PEX1 as the PBD group 1 gene (12), several PEX1 mutations were identified in group 1 patients. Of these, a substitution of aspartate for glycine at position 843 of the protein (G843D) was found to attenuate PEX1 activity in vivo, reducing its activity by approximately one-seventh. Significantly, the G843D mutation was found on 30% of PEX1 alleles in group 1 patients and is present in half of the CG1 patients, representing approximately one-fourth of all PBD patients and one-third of patients in the ZS spectrum. Thus, the G843D allele represents the single most common cause of the PBD. In this report, we present genetic and biochemical evidence for interaction between the peroxins PEX1 and PEX6. We also find that the PEX1 G843D mutation affects the PEX1–PEX6 interaction, suggesting that disruption of the PEX1–PEX6 interaction is the single most common cause of the PBD.
MATERIALS AND METHODS
Plasmids.
Plasmids designed to express PEX1 (pBER81) (12), PEX6 (pTY3) (10), PEX5 (14), PEX10 (Warren, unpublished material), and PEX12 (19) were derived from pcDNA3 (Invitrogen). The sequence GRSEQKLISEEDLNREQKLISEEDLNGEQKLISEEDLstop is encoded by the 3xmyc vector (pcDNA3–3xmyc) and lies between the BamHI and NotI sites of pcDNA3. PEX1–3xmyc carries this sequence at the 3′ end of the PEX1 ORF. The PEX1/G843D cDNA has been described (12), and the two-hybrid and 3xmyc expression vectors carrying this mutation were created by standard techniques (20). For two-hybrid studies we used the vectors pJL59 for expression of GAL4 DNA binding domain fusions and pPC86/L2 for expression of GAL4 transactivating domain fusions, derivatives of pPC62 and pPC86 (21) in which the yeast selectable markers TRP1 and LEU2 have been exchanged.
Cell Lines, Transfections, and Immunofluorescence.
Fibroblast cell lines of patients with PBD were cultured and transfected as described (6). Cells were processed for indirect immunofluorescence 2 days after transfection by sequential fixation, permeabilization, and antibody incubation by using accepted protocols (6). Anti-catalase antibodies were obtained from The Binding Site (San Diego) whereas Texas Red-conjugated secondary antibodies were obtained from Kirkegaard & Perry Laboratories. All micrographs were captured on an Olympus fluorescence microscope (Olympus, New Hyde Park, NY).
Relative rescue activities are expressed as the quotient of the differences in observed rescue between the cDNA of interest and vector alone, and the complementing cDNA and vector alone. Observed rescue is defined as the proportion of cells importing catalase in each population of transfected cells. In experiments with PBD108 cells, transfection with the PEX6 expression vector pTY3, the PEX1 expression vector pBER81, and the vector control (pcDNA3) yielded observed rescue values of 22% for pTY3 (440 cells importing catalase of 2,022 cells observed), 6.2% for pBER81 (128/2,052), and 2.2% for pcDNA3 (45/2,056). The values in the second trial were 23% (478/2,070) for pTY3, 7.0% (140/1,997) for pBER81, and 2.0% (40/2,011) for pcDNA3 and in the third trial were 25% (508/2,019) for pTY3, 7.0% (142/2,044) for pBER81, and 2.2% for pcDNA3. For PBD118 cells, rescue was observed in 557 cells of 1,554 observed (36%) in the population transfected with pBER81, 212 of 2,100 (1.0 × 101%) for those transfected with pTY3, and 17 of 1,800 (0.94%) in cells transfected with pcDNA3. Similar values were observed in trials 2 and 3. For PBD102 cells, observed rescue was 21% (441/2,089) for pTY3, 9.0% (180/2,000) for pBER81, and 5.6% (114/2,031) for pcDNA3. Similar values were observed in trials 2 and 3. Data was collected by indirect immunofluorescence and visual inspection of the cells, and import was scored when a cell showed punctate staining for catalase.
Two-Hybrid Analysis.
The two-hybrid reporter strain MaV99 was used for all experiments (22). Each strain was grown on selective medium (SC -trp, -leu), was transferred to a nitrocellulose filter placed on a yeast extract/peptone/dextrose plate supplemented with adenine (1.0 g/L), was grown for 2 days, and was lysed by submersion in liquid nitrogen. Activity of the two-hybrid reporter gene β-galactosidase was assessed by placing the cells filter side down on paper saturated with 0.1% 5-bromo-4-chloro-3-indoyl β-d-galactopyranoside in 100 mM sodium phosphate (pH 7.2). The filters were dried and photographed after overnight color development.
In Vitro Translation and Coimmunoprecipitations.
The pcDNA3 expression plasmids contain a T7 promoter sequence upstream of all cDNAs. Plasmid-directed synthesis of proteins in vitro was performed by using the TNT rabbit reticulocyte lysate system (Promega), and radiolabeled proteins were generated by addition of [35S]-methionine to the lysate. For coimmunoprecipitation experiments, equal amounts of [35S]-PEX6 were combined with equal amounts of unlabeled PEX1, PEX1–3xmyc, or PEX1/G843D-3xmyc, respectively, protease inhibitors were added (0.2 mM phenylmethylsulfonyl fluoride, 25 μg/ml aprotinin and leupeptin), and the volume of each sample was adjusted to 1 ml with binding buffer (0.2% Triton X-100/150 mM sodium chloride/0.2 mg/ml sodium fluoride/10 mM ATP/10 mM EDTA/50 mM Tris⋅HCl, pH 7.4). After incubation at 4°C for 30 min, 20 μl of monoclonal anti-myc (1–9E10) (23) conjugated agarose beads (Santa Cruz Biotechnology) were added, and the suspension was incubated at 4°C for 2 hr. The anti-myc beads were collected by centrifugation (4500 × g), were washed 10 times with 1 ml of IP buffer (binding buffer with 0.1% sodium dodecylsulfate), and were resuspended in 50 μl of SDS/PAGE sample buffer. Equal amounts of each immunoprecipitate were separated by SDS/PAGE (7.5%), and radiolabeled proteins were detected by fluorography.
RESULTS
Genetic Evidence for Interaction Between Human PEX1 and PEX6.
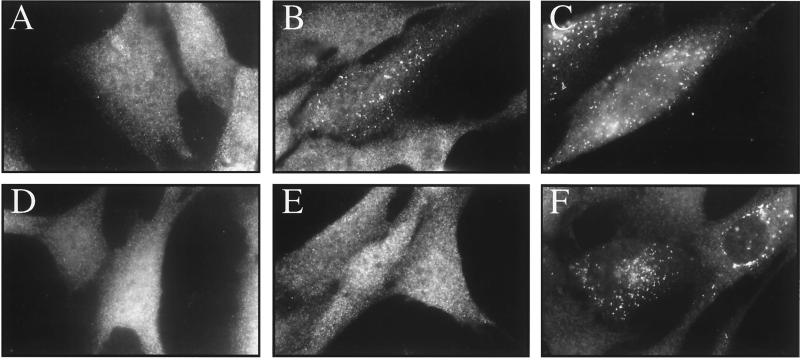
PBD group 1 cells have inactivating mutations in the PEX1 gene and are rescued phenotypically by transient expression of PEX1 (12, 17). However, during the course of routine controls, we made the rather surprising observation that overexpression of PEX6, the gene mutated in group 4 of the PBD, also rescued peroxisomal protein import in the PEX1-deficient cell line PBD118 (Fig. 1 A–C). In this particular assay, we examined the import of peroxisomal catalase in PBD118 cells that had been transfected with vector (pcDNA3), the PEX6 expression vector pTY3 (10), and the PEX1 expression vector pBER81 (12). Quantitation revealed that PEX6 was almost one-fourth as active as PEX1 in this assay, exhibiting 25% relative rescue as compared with 100% relative rescue for PEX1 (Table 1). These experiments were repeated three times, with relative rescue values of 23, 26, and 27% for PEX6. A critical control for these experiments was to test whether the punctate structures containing catalase were actually peroxisomes and to determine whether the catalase was enclosed within the peroxisome membrane. This was accomplished by performing double, indirect immunofluorescence experiments to localize catalase relative to a known peroxisomal protein, PMP70, and by differential permeabilization experiments, respectively. These controls were performed and confirmed that PEX6 overexpression did, indeed, result in catalase import into peroxisomes (data not shown). The suppression of PBD118 phenotypes was specific to PEX6 because overexpression of the human PEX5, PEX10, and PEX12 genes failed to restore peroxisomal protein import in PBD118 cells (data not shown).
Figure 1.
Expression of PEX6 rescues peroxisomal matrix protein import in PEX1-deficient group 1 cells in an allele-specific fashion. Human fibroblasts from a patient with NALD (PBD118; A–C) and a patient with ZS (PBD009; D–F) were transfected with pcDNA3 (A and C), derivatives of this plasmid designed to express the PEX6 (B and E), or PEX1 (C and F) cDNAs. Two days after transfection, the cells were processed for indirect immunofluorescence to determine the subcellular distribution of catalase, a PTS1-containing protein. Note that PEX6 expression mediated catalase import in the PBD118 cell line (B) but not in PBD009 cells (E) whereas transfection with the expression vector alone had no effect on catalase import in either line. Width of each image panel is 190 μm.
Table 1.
Relative rescue of PTS1-mediated peroxisomal protein import for individual cell lines expressing distinct PEX gene cDNAs
| Cell line | Gene mutated (phenotype) | Relative rescue activity of PEX gene cDNAs
|
||
|---|---|---|---|---|
| PEX1 | PEX6 | PEX1-3xmyc | ||
| PBD118 | PEX1 (NALD) | 100% | 25% | — |
| PBD009 | PEX1 (ZS) | 100% | 0 | 103% |
| PBD102 | PEX6 (NALD) | 22% | 100% | — |
| PBD108 | PEX6 (NALD) | 17% | 100% | — |
| PBD105 | PEX6 (ZS) | 1.0% | 100% | — |
Relative rescue was calculated as described in Materials and Methods.
PBD118 cells have different mutations on their two copies of PEX1. One allele contains a splice site mutation, resulting in aberrantly spliced PEX1 transcripts that are not expected to generate functional PEX1 protein (12). The other PEX1 allele of PBD118 cells carries the G843D mutation and has ≈15% of wild-type PEX1 activity, as determined by functional complementation. The presence of residual PEX1 activity was consistent with the mild clinical phenotypes of PBD118. The suppression of PBD118 cells suggested that PEX6 may either bypass the need for PEX1 or elevate residual PEX1 function through physical interaction. As a first test between these models, we examined the ability of PEX6 to rescue the peroxisomal protein import defects of a PEX1-deficient cell line that expresses no apparent PEX1 activity. PBD009 cells display the most severe peroxisomal protein import defects of any group 1 cell line and lack detectable PEX1 mRNA (12). We compared the relative rescue efficiencies of PEX1 and PEX6 in PBD009 cells and observed that PEX6 failed to rescue peroxisomal protein import in these cells (Fig. 1 D–F; Table 1). These results indicate that PEX6 is not able to bypass the need for PEX1 function and that the suppression of PBD118 cells by PEX6 requires at least some PEX1 activity.
To test the generality of genetic interaction between PEX1 and PEX6, we next examined the ability of PEX1 to suppress the peroxisomal protein import defects of PEX6-deficient fibroblast cell lines from CG4 of the PBD. Overexpression of PEX1 was able to restore peroxisomal protein import in PBD102 cells, though not as efficiently as PEX6 (Fig. 2 A–C; Table 1; relative rescue of 14, 22, and 15% in successive experiments), and also was able to suppress the import defects of the PEX6-deficient cell line PBD108 (Table 1; relative rescue of 21, 24, and 20% in successive experiments). Once again, import was confirmed by double labeling and differential permeabilization experiments (data not shown). The PEX6-deficient cell line PBD105 is a compound heterozygote for inactivating frameshift mutations in PEX6, lacks detectable PEX6 mRNA, and is derived from a severely affected patient with ZS. Overexpression of PEX1 could not suppress the import defects of this cell line (Fig. 2 D–F; Table 1), suggesting that suppression of PEX6 defects by overexpression of PEX1 is also allele-specific and requires residual PEX6 activity.
Figure 2.
Expression of PEX1 rescues PTS1-mediated import in selected PEX6-deficient cells. Human fibroblasts from a patient with NALD adrenoleukodystrophy (PBD102; A–C) and a patient with ZS (PBD105; D–F) were transfected with pcDNA3 (A and C), derivatives of this plasmid designed to express the PEX1 (B and E), or PEX6 (C and F) cDNAs. Two days after transfection, the cells were processed for indirect immunofluorescence to determine the subcellular distribution of catalase, a PTS1-containing protein. Note that PEX1 expression mediated catalase import in the PBD102 cell line (B) but not PBD105 cells (E) whereas transfection with the expression vector alone had no effect on catalase import in either line. Width of each image panel is 190 μm.
Physical Interaction Between the Products of PEX1 and PEX6.
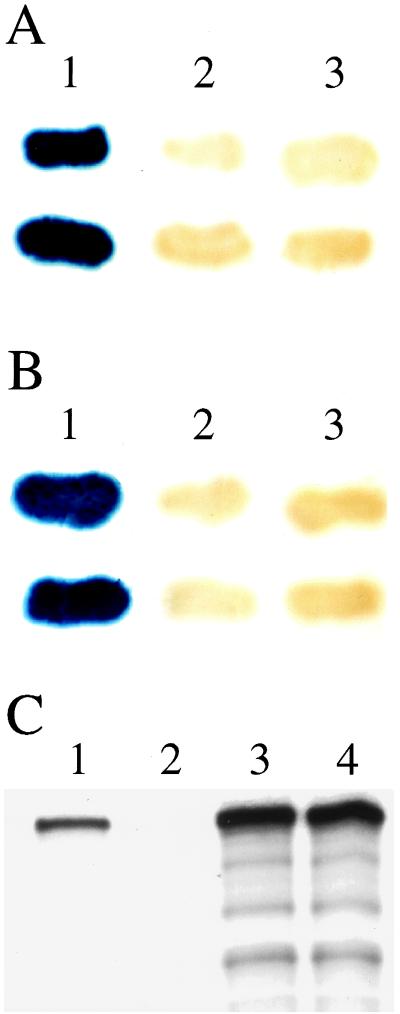
Allele-specific high-copy suppression is often indicative of direct physical interaction between the products of the interacting genes. We first tested this hypothesis for PEX1 and PEX6 by using the yeast two-hybrid system. Vectors that express human PEX1 fused to the GAL4 activating domain (G4AD–PEX1) and human PEX6 in fusion with the GAL4 DNA binding domain (G4BD–PEX6) were constructed. These were used to assess physical interaction between PEX1 and PEX6 by using a two-hybrid reporter strain, MaV99, which carries a chromosomal copy of a GAL4-regulated β-galactosidase reporter gene. GAL4 activity was observed in strains expressing G4AD–PEX1 and G4BD–PEX6, indicating physical interaction between PEX1 and PEX6 (Fig. 3A, lane 1). The fact that control strains expressing G4AD–PEX1 with G4BD (Fig. 3A, lane 2) or G4AD with G4BD–PEX6 (Fig. 3A, lane 3) failed to activate the GAL4-regulated reporter gene demonstrated that the reconstitution of GAL4 activity required coexpression of G4BD–PEX6 and G4AD–PEX1. Many aspects of peroxisomal protein import are conserved among eukaryotes, including yeast and humans. One approach to determining the general relevance of the putative PEX1–PEX6 interaction was to test whether it too might be an evolutionarily conserved feature of peroxisome biogenesis. Therefore, we assayed the Saccharomyces cerevisiae forms of PEX1 and PEX6. High levels of GAL4-regulated β-galactosidase activity were detected in MaV99 strains expressing both G4AD–ScPEX1 and G4BD–ScPEX6 (Fig. 3B, 1) but not in control strains expressing G4AD–ScPEX1 and G4BD alone (Fig. 3B, lane 2) or in control strains expressing G4BD–ScPEX6 and G4AD alone (Fig. 3B, lane 3). Thus, the PEX1–PEX6 interaction does appear to be a conserved feature of these proteins.
Figure 3.
Yeast two-hybrid and coimmunoprecipitation experiments confirm a physical interaction between the PEX1 and PEX6 proteins. Significant β-galactosidase activity was detected by filter assay of two-hybrid reporter strains coexpressing G4AD–PEX1 and G4BD–PEX6 (A, lane 1) but not in strains coexpressing either G4AD–PEX1 and G4BD (A, lane 2) or G4AD and G4BD–PEX6 (A, lane 3). Significant β-galactosidase activity also was detected in cell lysates of strains coexpressing S. cerevisiae forms of PEX1 and PEX6 (G4AD–ScPEX1 and G4BD–ScPEX6; B, lane 1) but not in strains coexpressing G4AD–ScPEX1 and G4BD (B, lane 2) or G4AD and G4BD–ScPEX6 (B, lane 3). In an independent set of experiments (C), cell free lysates containing [35S]-labeled PEX6 and either unlabeled PEX1–3xmyc or unlabeled PEX1 were subjected to immunoprecipitation with monoclonal anti-myc antibodies. The resulting immunoprecipitates were separated by SDS/PAGE and were assayed by fluorography, revealing significant amounts of [35S]-labeled PEX6 in the immunoprecipitate from the lysate containing PEX1–3xmyc (C, lane 1) but not in the immunoprecipitate from the lysate containing unmodified PEX1 (C, lane 2). Equal amounts of [35S]-labeled PEX6 in these two lysates before immunoprecipitation was confirmed by SDS/PAGE and fluorography (C, lanes 3 and 4, respectively). Control experiments confirmed that PEX1 and PEX1–3xmyc are synthesized equally in vitro and that only PEX1–3xmyc is immunoprecipitated with monoclonal anti-myc antibodies (data not shown).
Although these data indicate that PEX1 and PEX6 bind one another, the yeast two-hybrid system provides only an indirect measurement of physical interaction. We next examined the ability of human PEX1 and PEX6 to bind one another in a cell-free system and to remain associated during copurification. We synthesized [35S]-labeled PEX6 in vitro and combined the radiolabeled product with a myc-tagged form of PEX1, PEX1–3xmyc. This form of PEX1 contains three copies of the 10-aa c-myc epitope tag at its C terminus and can be immunoprecipitated specifically by using the mAb 1–9E10 (23). As a control for specificity, an equal amount of radiolabeled PEX6 was combined with unmodified PEX1. These two samples then were subjected to immunoprecipitation using the anti-myc mAb, separation by SDS/PAGE, and detection of labeled PEX6 by fluorography. The coimmunoprecipitation of PEX6 and PEX1–3xmyc indicates that PEX1 and PEX6 are capable of forming a stable complex in vitro (Fig. 3C, lane 1). The specificity of the coimmunoprecipitation for only the myc-tagged protein is demonstrated by the lack of PEX6 in the immunoprecipitate from the sample with unmodified PEX1(Fig. 3C, lane 2). The presence of equal amounts of PEX6 in the two samples before immunoprecipitation was confirmed by SDS/PAGE and fluorography (Fig. 3C, lanes 3 and 4) and the presence of equal amounts of PEX1 and PEX1–3xmyc in these samples also was confirmed (data not shown).
An assumption of these copurification experiments was that the addition of the 37-aa 3xmyc tag to the C terminus of PEX1 did not have a significant affect on its activity and, thus, that its behavior would resemble that of unmodified PEX1. We tested this hypothesis by examining the relative rescue activities of PEX1 and PEX1–3xmyc. PBD009 cells, which lack PEX1 mRNA and protein, were transfected with vector alone, the PEX1 expression vector pBER81, and an analogous plasmid designed to express PEX1–3xmyc. Rescue of peroxisomal protein import was detected by indirect immunofluorescence using antibodies specific for peroxisomal catalase and was quantitated by counting the number of cells in the population able to import catalase into peroxisomes. The relative rescue activity of PEX1–3xmyc was 103%, as compared with 100% for PEX1 (Table 1). We conclude that addition of the 3xmyc tag did not have a deleterious affect on PEX1 activity.
The G843D Mutation Attenuates the Interaction Between PEX1 and PEX6.
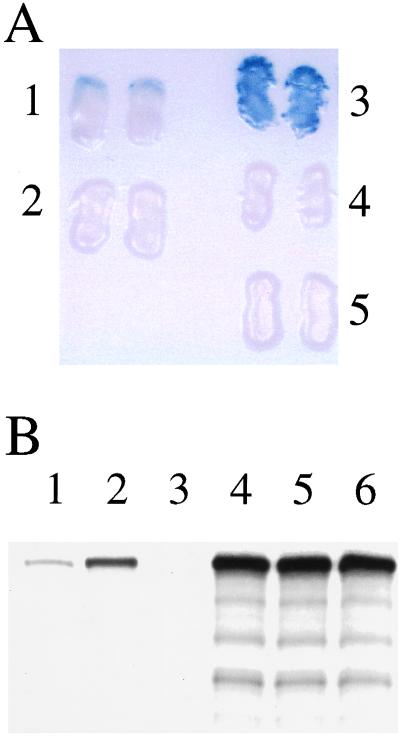
Half of all CG1 patients carry the G843D PEX1 allele, and this mutation reduces PEX1 activity by approximately one-seventh, as determined by in vivo functional complementation (12). Because overexpression of PEX6 suppressed the phenotypes of PBD118 cells and PBD118 cells carry the G843D mutation on one of their PEX1 alleles (Fig. 1), we hypothesized that the G843D mutation might attenuate the PEX1–PEX6 interaction. This was tested first in the two-hybrid system. The G843D mutation was inserted into the G4AD–PEX1 plasmid to create G4AD–PEX1/G843D. Derivatives of the two-hybrid reporter strain expressing G4BD–PEX6 and either G4AD-PEX1 or G4AD-PEX1/G843D were assayed for GAL4-regulated β-galactosidase activity. As evident from filter assays, the β-galactosidase activity exhibited by strains expressing G4AD–PEX1/G843D and G4BD–PEX6 (Fig. 4A, lane 1) was lower than the activity of strains expressing G4AD–PEX1 and G4BD–PEX6 (Fig. 4A, lane 3), suggesting that this mutation may indeed affect the PEX1–PEX6 interaction. If true, this mutation also should affect the ability of these proteins to form a stable complex in vitro. To test this possibility, we synthesized [35S]-labeled PEX6 and examined its binding to PEX1/G843D. Equal amounts of radiolabeled PEX6 were added to lysates containing equal amounts of either PEX1–3xmyc or PEX1/G843D-3xmyc. Each sample then was subjected to immunoprecipitation using mAb to the myc epitope tag, separation of the precipitate by SDS/PAGE, and quantitation of PEX6 levels in each precipitate by fluorography. The amount of PEX6 bound by the mutant form of PEX1 (Fig. 4B, lane 1) was only 30% of that bound by wild-type PEX1 (Fig. 4B, lane 2), providing direct evidence that the G843D mutation affects the PEX1–PEX6 interaction.
Figure 4.
The high-frequency G843D mutation in PEX1 attenuates the interaction between PEX1 and PEX6. Two-hybrid reporter strains coexpressing G4BD–PEX6 and G4AD–PEX1/G843D (A, lane 1) displayed significantly less β-galactosidase activity by filter assay than strains coexpressing G4BD–PEX6 and G4AD–PEX1 (A, lane 3). Control strains expressing G4BD and G4AD–PEX1/G843D (A, lane 2), G4BD and G4AD–PEX1 (A, lane 4), or G4BD–PEX6 and G4AD (A, lane 5) did not display detectable β-galactosidase activity. Similarly, SDS/PAGE and fluorography (B) revealed that anti-myc immunoprecipitates from lysates containing [35S]-labeled PEX6 and PEX1/G843D-3xmyc (B, lane 1) contained 70% less PEX6 than immunoprecipitates from lysates containing [35S]-labeled PEX6 and PEX1–3xmyc (B, lane 2). Once again, PEX6 was not detected in the anti-myc immunoprecipitate of lysates containing [35S]-labeled PEX6 and untagged PEX1 (B, lane 3). Equal amounts of PEX6 (B, lanes 4–6, respectively) were present in these three lysates before immunoprecipitation with anti-myc antibodies.
DISCUSSION
Peroxisome biogenesis requires the concerted action of multiple proteins, two of which are PEX1 and PEX6. In this study, we observed interaction between PEX1 and PEX6 in the two-hybrid system, coimmunoprecipitation of PEX1 and PEX6 from cell-free lysates, and allele-specific suppression of mutations in PEX1 or PEX6 by overexpression of PEX6 or PEX1, respectively. Together, these results provide both biochemical and genetic evidence that PEX1 and PEX6 interact with one another and that this interaction is biologically significant. Similar results have been observed independently for the P. pastoris forms of PEX1 and PEX6 (24). A logical deduction from these data is that PEX1 and PEX6 participate in a common aspect of peroxisome biogenesis. If so, loss of either protein would be expected to cause similar phenotypes. In human cells, loss of PEX1 results in reduced import of PTS1 and PTS2 proteins and a pronounced decrease in the stability and abundance of PEX5 (11); however, these same cells display normal import of PMPs and contain ≈100 PMP-containing peroxisomes per cell (Chang and S.J.G., unpublished data). These phenotypes are almost identical to those observed for group 4 PBD cells lacking PEX6 (10). Thus, it appears that PEX1 and PEX6 are required for common aspects of peroxisome biogenesis. The significance of congruent pex1 and pex6 phenotypes is underscored by the distinct phenotypes of cells lacking other peroxins involved in PTS1 and PTS2 protein import. For instance, severe mutations in the human PEX2, PEX10, or PEX12 genes (in PBD groups 10, 7, and 3, respectively) block matrix protein import completely but have no effect on the stability of PEX5 (11). In fact, the only other PBD cells that display phenotypes similar to those of pex1 or pex6 mutants are from CG8. Although the interaction between PEX1 and PEX6 in multiple species suggests conserved features to their roles in peroxisome biogenesis, there may be significant differences in the phenotypes of pex1 and pex6 mutants in different organisms. Most notably, studies in the yeast Yarrowia lipolytica have demonstrated that these proteins play an essential role in protein secretion (25), a phenotype that is not apparent in the corresponding human mutants.
Because the PEX1–PEX6 interaction is important for peroxisome biogenesis, mutations that affect this interaction would be expected to result in human disease. The G843D mutation in PEX1 is the single most common cause of the PBD and is present in one-half of all PEX1-deficient patients and one-third of all patients in the ZS spectrum (12). The original suppression of a PEX1-deficient cell line by PEX6 overexpression was observed in PBD118 cells that have one copy of the G843D allele. The G843D mutation does not inactivate PEX1 but reduces its activity by >80% (12). We observed that the G843D mutation reduces the amount of PEX6 bound to PEX1 by ≈70% (as observed in copurification experiments) and attenuates the PEX1–PEX6 interaction in the two-hybrid system. Thus, it appears that disruption of the PEX1–PEX6 complex represents the single most common cause of the PBD. Although our data for the human PEX1–PEX6 complex suggests a specific role in peroxisomal matrix protein import, we currently are testing this hypothesis more rigorously.
Acknowledgments
This work was supported by grants from the National Institutes of Health to S.J.G. (DK45787 and HD10891).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PBD, peroxisome biogenesis disorders; PTS, peroxisome targeting signal; ZS, Zellweger syndrome; NALD, neonatal adrenoleukodystrophy CG, complementary group; AD, activating domain; BD, binding domain.
References
- 1.Lazarow P B, Fujiki Y. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 2.Subramani S. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- 3.Gould S J, Keller G A, Hosken N, Wilkinson J, Subramani S. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swinkels B W, Gould S J, Bodnar A G, Rachubinski R A, Subramani S. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazarow P B, Moser H W. In: Disorders of Peroxisome Biogenesis. Scriver C, Beaudet A, Sly W, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2287–2324. [Google Scholar]
- 6.Slawecki M, Dodt G, Steinberg S, Moser A B, Moser H W, Gould S J. J Cell Sci. 1995;108:1817–1829. doi: 10.1242/jcs.108.5.1817. [DOI] [PubMed] [Google Scholar]
- 7.Motley A, Hettema E, Distel B, Tabak H. J Cell Biol. 1994;125:755–767. doi: 10.1083/jcb.125.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimozawa N, Suzuki Y, Orii T, Moser A, Moser H W, Wanders R J A. Am J Hum Genet. 1993;52:843–844. [PMC free article] [PubMed] [Google Scholar]
- 9.Distel B, Erdmann R, Gould S J, Blobel G, Crane D I, Cregg J M, Dodt G, Fujiki Y, Goodman J M, Just W W, et al. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yahraus T, Braverman N, Dodt G, Kalish J E, Morrell J C, Moser H W, Valle D, Gould S J. EMBO J. 1996;15:2914–2923. [PMC free article] [PubMed] [Google Scholar]
- 11.Dodt G, Gould S J. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuber B E, Germain-Lee E, Collins C S, Morrell J C, Ameritunga R, Moser H W, Valle D, Gould S J. Nat Genet. 1997;17:445–448. doi: 10.1038/ng1297-445. [DOI] [PubMed] [Google Scholar]
- 13.Gould S J, Keller G A, Schneider M, Howell S H, Garrard L J, Goodman J M, Distel B, Tabak H, Subramani S. EMBO J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodt G, Braverman N, Wong C, Moser A, Moser H W, Watkins P, Valle D, Gould S J. Nat Genet. 1995;9:115–124. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- 15.Dodt, G., Braverman, N., Valle, D. & Gould, S. J. (1998) Ann. N. Y. Acad. Sci. 804, in press. [DOI] [PubMed]
- 16.Fukuda S, Shimozawa N, Suzuki Y, Zhang Z, Tomatsu S, Tsukamoto T, Hashiguchi N, Osumi T, Masuno M, Imaizumi K, et al. Am J Hum Genet. 1996;59:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 17.Portsteffen H, Beyer A, Becker E, Epplen C, Pawlak A, Kunau W-H, Dodt G. Nat Genet. 1997;17:449–452. doi: 10.1038/ng1297-449. [DOI] [PubMed] [Google Scholar]
- 18.Moser A, Rasmussen M, Naidu S, Watkins P, McGuinness M, Hajra A, Chen G, Raymond G, Liu A, Gordon D, et al. J Pediatr. 1995;127:13–22. doi: 10.1016/s0022-3476(95)70250-4. [DOI] [PubMed] [Google Scholar]
- 19.Chang C-C, Lee W-H, Moser H W, Valle D, Gould S J. Nat Genet. 1997;15:385–388. doi: 10.1038/ng0497-385. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Chevray P M, Nathans D. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal M, Brachman R K, Fattaey A, Harlow E, Boeke J D. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evan G E, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faber K N, Hetman J A, Subramani S. Mol Cell Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Titorenko V I, Ogrydziak D M, Rachubinski R A. Mol Cell Biol. 1997;17:5210–5226. doi: 10.1128/mcb.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]