Abstract
The perinuclear localization of myosin-V was investigated in a variety of cultured mammalian cells and in primary cultures of rat hippocampus. In all cells investigated, myosin-V immunoreactivity was associated with the centrosome. In interphase cells, myosin-V was found in pericentriolar material, and in both mother and daughter centrioles. These results were obtained by using two different fixation protocols with three different affinity-purified antibodies that recognized a single band in Western blots. During cell division, myosin-V staining was intense throughout the cytoplasm and was concentrated in a trail between migrating centrioles and in the mitotic spindle poles and spindle fibers. The centrosome targeting site was determined to reside within the globular tail domain, because centrosome association also was observed in living cells transfected with DNA encoding the tail domain fused with a green fluorescent protein tag, but not in cells transfected with the vector encoding green fluorescent protein by itself.
Keywords: mitotic spindle
The ability to move and position organelles is essential to eukaryotic cellular physiology. Motility is accomplished by mechanochemical ATPases, dyneins, and kinesins that translocate on microtubules, and myosins that translocate on actin filaments. At least 15 classes of myosins have been identified (1, 2).
The range of functions of most of the novel myosins have yet to be fully characterized. Myosin-V is a dimeric, nonfilamentous myosin with calmodulin light chains and actin-dependent ATPase activity showing an absolute requirement for calcium (3, 4). Class V myosins, for which full-length sequences have been determined, include the protein products of the mouse Myo5a gene (dilute locus) (5) and the yeast MYO2 (6) and MYO4 (7) genes, chicken myosin-Va (8, 9), human myosin-Va (MYH12, GenBank accession no. Y07759), and rat myosin-Vb (myr6) (10).
Phenotypic analyses of yeast myo2 and myo4 and mouse dilute mutants have shown that myosin-V is required for the polarized transport of organelles and other cytoplasmic components. Myo2–66 yeast cells, which express a temperature-sensitive mutation of a myosin-V gene (6), cannot progress through the cell cycle at restrictive temperatures, but remain as large, unbudded cells with numerous small vesicles in the cytoplasm. The defective gene, Myo2p, is required for vacuole inheritance and for the polarized localization of chitin synthase Chs3p and SEC4 in the budding yeast. Myo4p, another isoform of myosin-V in yeast, is required for the restricted localization of ASH1 mRNA to the daughter cell. As a consequence, Ash1p, a repressor of mating type switching, is selectively expressed in the daughter cell (for review see ref. 1).
Mutation of the mouse Myo5a gene results in defective pigment granule transfer (11) and, in such homozygous mutants as dl20J (dilute-lethal), may lead to neurological impairment that culminates in early postnatal death (5). Lack of smooth endoplasmic reticulum in the dendritic spines of Purkinje cells has been demonstrated in this mouse mutant (12) and in an equivalent rat mutant (13). In neurons and glial cells, myosin-V staining was found to be punctate and abundant in the perinuclear region, along the cellular processes and in the distal tips of processes (8, 14). Recent studies have shown myosin-V to be associated with synaptic vesicles and vesicles colocalized with microtubules and F-actin in neuronal growth cones, as well as to be involved in filopodial extension (for review see refs. 1 and 15). Consistent with a role in vesicular transport, direct association of myosin-V with small vesicles, melanosomes at all stages of maturation, and endoplasmic reticulum has been shown by immunogold staining in melanoma cells (16). By light microscopy, myosin-V staining in melanoma cells was shown to be punctate, prominent in the perinuclear region, and colocalized with melanosomes (11, 16, 17). Together, these findings indicate that class V myosins perform multiple functions within the cell.
In this study, we used two immunocytochemical fixation protocols and three affinity-purified antibodies as well as transfection of DNA encoding the myosin-Va globular tail domain fused with a green fluorescent protein (GFP) tag to characterize myosin-V localization. We employed a variety of highly proliferating cell lines, as well as primary cultures of brain tissue. We report a consistent association of myosin-V with the centrosome, at all stages of the cell cycle, by all methods used.
MATERIALS AND METHODS
Cells.
MDCK cells, a canine kidney cell line, were obtained from ATCC (Manassas, VA). The cells were grown in Eagle’s minimum essential medium with Earl’s salts and glutamine (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) at 37°C in 5% CO2.
Clonal cell lines from the organ of Corti (OC-k3) and stria vascularis (SV-k1) were isolated in this laboratory (F. Kalinec, unpublished data) from a transgenic mouse (Immortomouse) that constitutively expresses a temperature-sensitive simian virus 40 T antigen. The cells were grown at 33°C in 10% CO2 in DMEM (GIBCO), supplemented with 10% fetal bovine serum and 50 units/ml of recombinant mouse interferon γ (Genzyme) to induce the transgene. Under these conditions, cells proliferate very fast, doubling in number daily.
The B16-F10 murine melanoma cell line (derived from C57BL/6J mouse, D/D; ref. 18) was acquired as a generous gift from J. Pawelek (Yale University, New Haven, CT). Cells were grown in Ham-F10 (GIBCO), supplemented with 10% horse serum, at 37°C in 5% CO2. Under these conditions, cells proliferate very fast and show a low degree of pigmentation. To induce differentiation, the culture medium was changed to DMEM, made with 0.398 mM l-tyrosine, and supplemented with 2% fetal bovine serum plus 0.4 μM α-melanocyte stimulating hormone (α-MSH, Sigma) and 0.5 mM 3-isobutyl-1-methyl-xanthine (Sigma) for 1–2 days. Under these conditions, cells proliferate at a slower rate and become highly pigmented, flat, and dendritic.
S91–6 cells (19), a clone whose melanization and branching are dependent on α-MSH stimulus, are derived from the S91 murine melanoma cell line (isolated from a DBA/2J mouse dv/dv, b/b). These cells, also obtained from J. Pawelek, Yale University, were grown in Ham-F10 (GIBCO) supplemented with 10% horse serum, at 37°C in 5% CO2. Under these conditions cells were amelanotic and adendritic.
Primary cultures from rat hippocampus, a gift from D. Murphy (National Institute of Neurological Disorders and Stroke), were cultured as described elsewhere (20).
Antibodies.
Antitail polyclonal antibody, generated against a chicken myosin-V recombinant protein, has been characterized previously (8). The antigen used to produce this antibody consisted of a recombinant protein, expressed in bacteria, containing most of the tail domain of chicken myosin-Va. Affinity purification was achieved with a recombinant tail protein fused with maltose-binding protein. Two polyclonal antibodies were generated, independently, against a recombinant protein containing the myosin-V head domain (amino acids 5–752) in fusion with a maltose-binding protein. One antihead antibody, Hc, was generated against a soluble fusion protein purified on an amylose column and affinity-purified against chicken brain myosin-V immobilized on nitrocellulose. This antibody was shown previously to inhibit vesicle motility in in vitro assays by Wolenski et al. (21). The other antihead antibody, Hf (a gift from F. Espindola and M. Mooseker, Yale University), was generated against the same head antigen, except that the fusion protein was isolated by SDS/PAGE, followed by electroelution. Affinity purification was done in two steps, by chromatographic adsorption of antibodies against a maltose-binding protein, followed by affinity purification against the antigen coupled to a column. A mouse anti-β-tubulin mAb was obtained from Amersham. Secondary antibodies (fluorescein isothiocyanate-conjugated donkey anti-rabbit Ig and Texas Red-conjugated sheep anti-mouse Ig) were obtained from Amersham.
Immunocytochemistry.
Cells were fixed and permeabilized by using one of the following two protocols: (i) 2% paraformaldehyde in PBS containing 0.3% Triton X-100, for 10 min at 37°C, followed by washing three times with PBS at room temperature; and (ii) 1% paraformaldehyde in methyl alcohol for 10 min at −20°C, followed by washing three times with PBS. After fixation, cells were incubated for 1 h at room temperature in 2% BSA and 5% goat serum in PBS, then incubated at room temperature for 1–3 h with the primary antibody diluted in blocking solution (for anti-β-tubulin, 1:100; anti-myosin-V tail, 5 μg/ml; anti-myosin-V head Hc and Hf, 3 μg/ml). After incubation with the primary antibody, cells were washed five times with PBS, incubated for 1 h at room temperature with the secondary antibody (1:100 in blocking solution), and then washed five times with PBS. For double labeling of myosin-V and tubulin, incubations were done simultaneously. Coverslips were mounted in 1 mg/ml p-phenylenediamine (Sigma) in 90% glycerol made with PBS. Fluorescence label was observed by using a Zeiss Axiophot microscope equipped with ×63 and ×100, 1.4 numerical aperture, objectives (Zeiss) and recorded with a 35-mm photographic camera or a Photometrics cooled charge-coupled device (CCD) camera (Tucson, AZ).
Preparation of Total Homogenates from Cultured Cells for Electrophoresis and Immunoblotting.
MDCK, B16-F10, S91–6, and SV-k1 cells were grown as specified above, in nondifferentiating conditions, in 150-mm-diameter culture plates. When confluent, cells were scraped from the plates in 200 ml ice-cold buffer [40 mM Hepes, pH 7.7/10 mM EDTA/2 mM DTT/1 μM benzamidine/2 μg/ml aprotinin/1 mM pefabloc (Boehringer Mannheim)] and lysed with a Dounce-type glass homogenizer. Proteins from whole-cell lysates were separated by SDS/PAGE on 6–15% gradient gels, transferred to nitrocellulose, and probed with affinity-purified antibodies specific for the tail or head (Hf) of myosin-V, at 0.6 μg/ml and 0.1 μg/ml, respectively. Bound antibodies were detected by incubation with a peroxidase-conjugated anti-rabbit IgG and visualized by chemiluminescence (ECL, Amersham).
Subcloning.
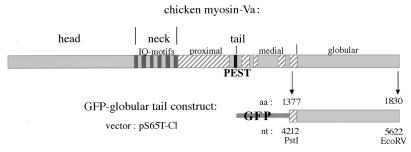
DNA encoding the myosin-Va globular tail was subcloned in fusion with GFP in the vector pS65T-C1 obtained from CLONTECH. This construct was generated by using the following strategy (Fig. 7). A DNA fragment insert corresponding to the globular tail of chicken myosin-Va [nucleotide position 4,212 (PstI site) to 5,622 (EcoRV site)] had been subcloned previously in the bacterial expression vector pTrc99A, from the original 32a clone in p-Bluescript (8), into the restriction sites PstI to HindIII. For the present subcloning, this fragment was cut from the pTrc99A vector with the restriction enzyme HindIII, blunted with Klenow fragment DNA polymerase, followed by a second restriction digestion with KpnI. This fragment was isolated and ligated into KpnI and SmaI sites of the pS65T-C1 vector in fusion with the GFP ORF so that the GFP is located at the amino terminus of the protein. The regions spanning the sites used for subcloning at both ends of the inserted DNA were sequenced and confirmed to contain the expected nucleotide sequence.
Figure 7.
Cloning strategy. Schematic diagram representing the linear domain structure of chicken myosin-Va and the GFP–globular tail fusion protein expressed from the construct obtained in the pS65T-C1 vector. The construct includes the full myosin-Va globular tail plus 45 aa residues from the medial tail as represented in the diagram, where the first and last amino acid and nucleotide (nt) positions are indicated.
Transfection.
S91 or B16 cells were grown in the appropriate culture medium until confluence, harvested in Tyrode solution (0.4 mM NaH2PO4/11.9 mM NaHCO3, pH 7.2/5.5 mM glucose/137 mM NaCl/2.7 mM KCl/0.68 mM EDTA), centrifuged, and resuspended in 400 μl of RPMI 1640 culture medium (GIBCO) to give approximately 107 cells/ml. Cells were transferred to sterile electroporation cuvettes (4-mm gap, BTX, San Diego) and mixed with 10 μg of purified plasmid DNA. Electroporation was performed in a BTX ECM-395 electroporator at 275 V. After electroporation cells were maintained on ice for 10 min and then plated in normal culture medium, observed after 48 h by using a Zeiss Axiovert microscope equipped with a ×63, 1.4 numerical aperture objective, and recorded with a Pentamax cooled CCD camera (Princeton Instruments, Trenton, NJ).
RESULTS
Centrosome Labeling of Interphase Cells.
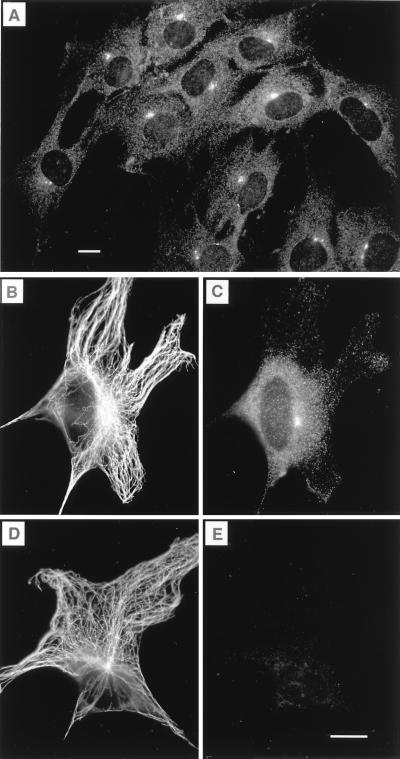
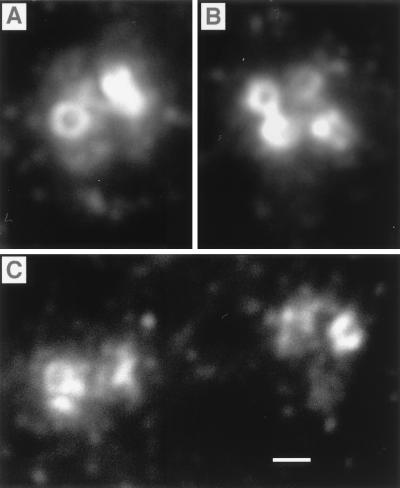
In interphase cells, cytoplasmic staining varied with cell type, but a common feature of myosin-V labeling was the presence of one or two foci that coincided with the microtubule organizing center (Fig. 1). The centrosome was labeled in all cells examined. Centrosome labeling was seen with each of three affinity-purified antibodies to myosin-V, including a highly specific antibody against the tail region of the molecule (8) and two antibodies independently raised against the head region. Labeling was not a result of secondary antibodies and could be blocked by soluble myosin-V purified from chicken brain (Fig. 1E). The concentration of myosin-V used for blocking did not affect the staining by antibodies against β-tubulin (Fig. 1 B and D). High magnification revealed a cluster of labeled spots surrounding the uniformly and intensely labeled centrioles (Fig. 2), suggesting that myosin-V is present on the surface of the centriole and is a component of the pericentriolar material, possibly in the pericentriolar satellite structures (22). Mother and daughter centrioles were labeled equally. Myosin-V was found on new centrioles after replication at the beginning of mitosis.
Figure 1.
Labeling of centrosomes with myosin-V antitail domain antibody in OC-k3 cells. (A) A field showing several interphase cells with intensely labeled foci in the perinuclear region. The three cells at the bottom appear to be in prophase (see Figs. 3 and 4). (B and C) A cell double-labeled with anti-β-tubulin and anti-myosin-V, respectively. The microtubule-organizing center coincided with the bright spot labeled with anti-myosin-V. (D and E) Preadsorption control cell labeled with anti-β-tubulin and anti-myosin-V plus 0.5 mg/ml chicken brain myosin-V, respectively. Preincubation with myosin-V did not affect the labeling by the anti-tubulin antibody, but completely blocked myosin-V labeling. Cells were fixed by using method ii (Materials and Methods). (Bars = 10 μm.)
Figure 2.
High magnification of centrosomes labeled with the anti-tail antibody. (A) A pair of centrioles in an interphase cell. (B) Two pairs after replication. (C) Two pairs in early prophase just after poleward migration has begun. In each centrosome, the entire surface of the centrioles and a cluster of surrounding punctae were labeled. Cells were fixed by using method i (Materials and Methods). Centrosomes in A and B are from SV-k1 cells. Those in C are from B16-F10 cells. (Bar = 0.5 μm.)
Distribution during mitosis.
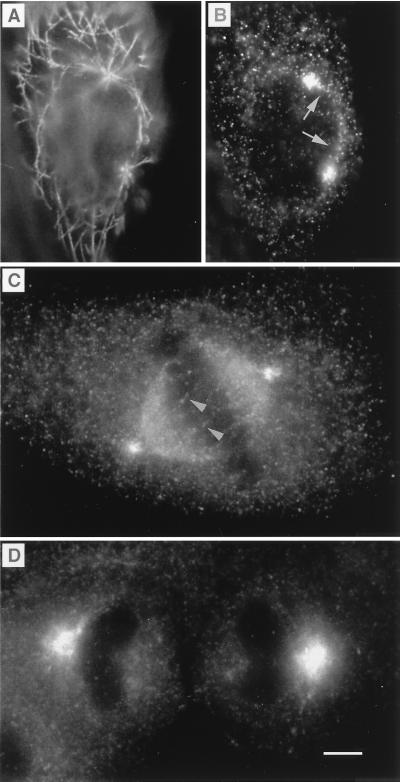
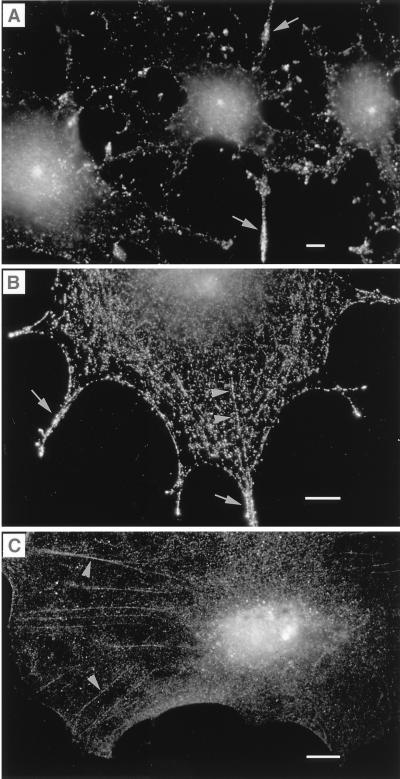
During mitosis, myosin-V labeling increased. Myosin-V was detected, not only in the centrosome and spindle poles, but also in a diffuse manner throughout the cytoplasm (Figs. 3 and 4). Increased expression was found in all cell types analyzed and with all three myosin-V antibodies. In prophase, myosin-V was concentrated at both centrosomes and along the line that separates the two migrating pairs (Fig. 3 A and B). At metaphase, anti-myosin-V labeled spindle poles and fibers (Fig. 3C) but not aster microtubules. Spindle labeling was punctate, and some fibers with punctate label were observed crossing the metaphase plate (Fig. 3C), suggestive of polar microtubules. The increased intensity of myosin-V labeling became noticeable during metaphase and persisted throughout anaphase and telophase (Fig. 3D).
Figure 3.
Labeling of MDCK cells at different stages of the cell cycle. During prophase, the migrating microtubule-organizing centers were labeled with antitubulin (A), which coincided, at the poles, with myosin-V labeling (B). A trail of myosin-V labeling typically could be visualized running along the rim of the nucleus on the path of the migrating centrioles (arrow). At metaphase (C), myosin-V labeling could be visualized on spindle fibers and as a diffuse labeling throughout the spindle. To visualize detail in the regions of the poles and spindle, in metaphase cells (C) and in cells progressing from anaphase to telophase (D), brightness and contrast have been adjusted, so that the pronounced increase in diffuse cytoplasmic staining (text and Fig. 4) is not as obvious. Cells were fixed by using method i (Materials and Methods). Myosin-V antibodies were antitail (B), antihead, Hc (C), and antihead, Hf (D). (Bar = 5 μm.)
Figure 4.
Double-labeling of cells with antibodies against myosin-V (green) and β-tubulin (red), showing the dramatic increase in the intensity of myosin-V staining in mitotic cells. The yellow color results from an overlap of the green and red. (A) Several stages of the cell cycle can be seen in SV-k1 cells, including interphase (most cells), prophase (arrow), and metaphase (intensely stained cells near the center). Higher magnification of metaphase cells (B, OC-k3 cells and C, SV-k1 cells) shows myosin-V labeling in the spindle and poles, and an intense, diffuse label throughout the cytoplasm. Cells were fixed by using method ii (Materials and Methods). Myosin-V antibodies were antitail (A and B) and antihead, Hf (C). (Bars = 10 μm.)
No indication of association of myosin-V with the kinetochore or centromere was observed. During and after metaphase, a diffuse cytoplasmic labeling filled the entire cytoplasm (Fig. 4), suggesting that there is an increase in the pool of free myosin-V.
Centrosome Localization in Differentiated Cells.
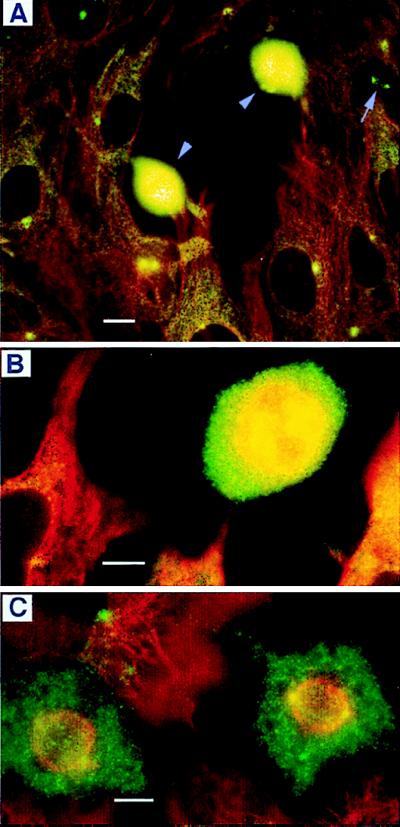
Myosin-V also was found in the centrosome of differentiated cells. B16-F10 melanoma cells (Fig. 4C) divide more slowly and become pigmented and dendritic (Fig. 5 A and B) when induced to differentiate. As the cells differentiated, myosin-V staining was abundant in the perinuclear region, always more intense in a perinuclear spot suggestive of the centrosome. Labeling also was distributed throughout the cytoplasm and dendrites in a punctate pattern. In the cytoplasm, punctae frequently appeared to be aligned along stress fibers (Fig. 5 A and B). This cellular distribution suggests multiple cellular functions for this mechanoenzyme.
Figure 5.
Myosin-V labeling in differentiated cells. (A and B) B-16 cells induced to differentiate. (C) A rat hippocampus glial cell from a 3-week-old primary culture. Cells were labeled on the periphery, along stress fibers (arrowheads) and in the dendritic processes (arrows) in addition to the bright labeling of the centrosome. Cells were fixed by using method i (Materials and Methods) and labeled with myosin-V antihead antibody, Hc. (Bars = 10 μm.)
Centrosome localization was not restricted to immortalized cell lines. Myosin-V was also detected at the centrosome in primary cultures of glial cells (Fig. 5C) and neurons (data not shown) from mammalian brain.
Myosin-V Tail- and Head-Specific Antibodies Recognize a Single Band in Western Blots.
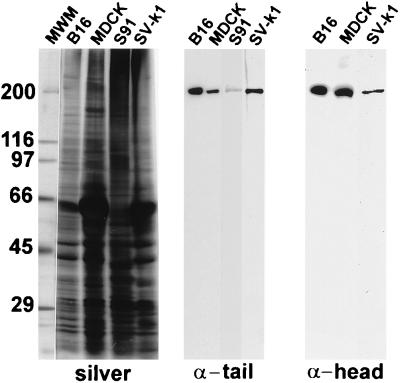
Western blots containing total homogenates of these cells were probed with myosin-V tail- and head-specific antibodies. In MDCK, B16-F10, SV-k1, and S91 cells, antibodies against both domains recognized a single, 190-kDa band expected for myosin-V (Fig. 6).
Figure 6.
Antibodies raised against myosin-Va tail and head domains are specific for a 190-kDa band in Western blots of total homogenates of several cell lines. Silver-stained gel containing molecular weight markers (MWM) and total homogenates of B16-F10, MDCK, S91, and SV-k1 cells and Western blots containing equivalent amounts of the specified homogenates probed with the antitail and antihead Hf antibodies. Both antibodies recognize a single band migrating at approximately 190 kDa in SDS/PAGE, as expected for myosin-V.
Myosin-Va Globular Tail Targets the Centrosome.
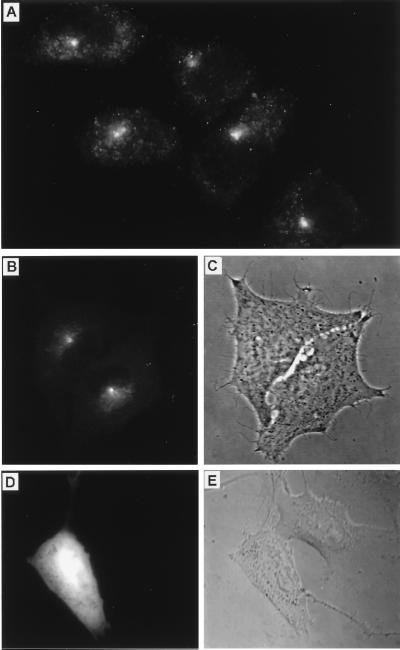
To further dissect the distribution of myosin-V in specific cytoplasmic domains in mammalian cells, a construct (Fig. 7) was generated with which myosin-V globular tail domain fused with GFP could be expressed in living cells. In initial experiments, we transfected S91 cells. Myosin-V is expressed in low levels in these cells. The antibody labeling was concentrated at the centrosome in proliferating (nonstimulated) cells (Fig. 8A). The main localization of the globular tail–GFP fusion protein was also the centrosome (Fig. 8 B and C). Centrosome localization of the globular tail–GFP fusion protein was detected not long after transfection (at low levels of protein expression) in all cells tested. Transfection with the original vector expressing GFP alone resulted in a diffuse localization of the fluorescent tag throughout the cytoplasm and nucleus of cells (Fig. 8 D and E). Transfection with other myosin-V structural domains fused with GFP resulted in distinct localizations for each specific domain (unpublished work).
Figure 8.
The globular tail domain of myosin-Va is targeted to the centrosome in mammalian cells. (A) Immunofluorescence staining for myosin-V in interphase S91 cells using the antitail antibody. (B–E) Live, transfected cells imaged by fluorescence microscopy using a filter set for fluorescein isothiocyanate (B and D) and phase-contrast microscopy (C) or differential interference contrast microscopy (E) of the same fields, respectively. (B and C) Recently divided S91 cells transfected with the GFP–globular tail construct. (D and E) B16-F10 cells transfected with the pS65T-C1 control vector showing diffuse fluorescence from GFP alone.
DISCUSSION
In this study, myosin-V is identified as a centrosome-associated protein by stringent immunocytochemical analysis using three different, affinity-purified antibodies against two distinct structural domains. Targeting of the GFP fusion protein independently supports this identification and indicates that the centrosome targeting site resides in the myosin-Va globular tail domain.
Localization of myosin-V on spindle fibers and increased intensity of myosin-V labeling during mitosis are also intriguing. We interpret the increase in labeling intensity as increased protein expression. It is likely that the increased expression and the centrosome association are related to distinct functions of myosin-V during cell division.
Mitosis is a dynamic period of the cell cycle. In addition to the separation of chromosomes, it is characterized by the disassembly, partitioning, and reorganization of intracellular membranous organelles. Elevated expression of myosin-V during mitosis parallels an increase in the number of cellular vesicles that result from the breakdown of the nuclear envelope and of Golgi cisternae (23, 24) and with the suppression of both plus- and minus-end-directed vesicular traffic on microtubules (25, 26). Considering its apparent role in vesicular transport (1, 15, 21), myosin-V may have an important role during cytokinesis in the partitioning of vesicles and in the re-formation of membrane-bound organelles.
The centrosome, named for its central location in the perinuclear region of most eukaryotic cells, usually contains two centrioles surrounded by amorphous pericentriolar material. The centrosome is the principal microtubule-organizing center of the cell (27, 28). Centrosomes are complex structures composed of a large number of proteins of unknown molecular functions (29). Although the specific role of the myosin-V in centrosome function is unknown, its importance is underscored by its function as a mechanoenzyme and by its association with calcium and calcium-binding proteins. Calmodulin, a major calcium-binding regulatory protein, is the principal myosin-V light chain (3, 8, 30). Calmodulin (31) and centrin (32), a homologous protein, also have been localized to the centrosome and to the mitotic apparatus in vertebrate cells. A yeast homologue of centrin, cdc 31 (33, 34), is essential for duplication of the spindle pole body, the yeast analog of the centriole. Calcium signaling initiates several events in the cell cycle, in part through the activation of calmodulin-dependent protein kinase II (35). Myosin-V is a known substrate for this kinase in in vitro assays (36) and, thus, a potential effector of calcium-dependent mitotic events.
The S91 cell line used in this study is a useful model for the characterization of class V myosins in the centrosome because nearly all of the myosin-V labeling is localized in that organelle (Fig. 8A). The low levels of myosin-Va protein expression (Fig. 6) agree with low levels of mRNA expression (37). Further, we have not detected myosin-Vb, by reverse transcription–PCR or Western blot analyses (unpublished data). Thus, myosin-Va (Myo5a) gene product is likely the major centrosome-associated myosin-V isoform.
It is important to note that myosin-Va is not essential for cell division. Mice carrying a null mutation of the Myo5a gene, such as dl20J (5), may survive up to 3 weeks after birth. Homozygous dl20J pups are smaller than heterozygous littermates, but their general appearance indicates that cell division and organ differentiation have occurred in a near normal manner. This suggests that either subtle defects in centrosome structure or function await to be discovered or that another gene product is compensating for the loss of myosin-Va.
Our data do not address the precise function of myosin-V in the centrosome. However, it is important to consider observations that may point toward functional experiments. Melanocytes isolated from dilute lethal mice have been reported to divide at half the rate of melanocytes isolated from wild-type mice (11). Another intriguing observation is the recent discovery that the Gricelli disease is associated with mutations in the Myosin-Va gene (38). This disease is marked by a severe cellular immunodeficiency. The link with myosin-V leads to the speculation that myosin-Va function in the centrosome of human lymphocytes may be essential either for cellular proliferation or for the polarized movement of the centrosome that occurs during T killer or T helper cell response (39).
The study of centrosome dynamics in interphase and dividing cells has been focused mainly on microtubule-based motility (40, 41). There have been several reports supporting the participation of actin and myosin in centrosome-associated processes (41–46). However, the data available are insufficient to elucidate a precise role for actomyosin system in mitotic and other centrosome-associated functions. Although localization of the product of the MYO2 gene varies strikingly during the Saccharomyces cerevisiae cell cycle (47), there has been no indication of its localization in the spindle pole body. However, interesting clues for the interaction of myosin-V with microtubule-based motors are emerging. Overexpression of the kinesin-like SMY1p not only rescues the myo2–66 mutation, but also restores the wild-type localization of the mutant MYO2 gene product (48). In addition, interaction of myosin-V with the 10-kDa cytoplasmic dynein light chain has been demonstrated recently (49). This light chain also has been found at the centrosome (50).
Identification of myosin-V as a centrosome-associated protein suggests a number of experiments that should lead to a better understanding of centrosome function. The findings provide new possibilities for a concerted interaction between actin and microtubule-based motility and support the concept of multifunctional roles for class V myosins (15).
Acknowledgments
We thank M. C. R. Costa, A. A. C. Nascimento, S. R. Banzi, and S. B. F. Tauhata for the purification of myosin-V and myosin-V antibodies Hc; E. V. Patussi for help with the Western blots; and J. Fex, R. Petralia, and H. Arnheiter for critical comments on the manuscript. The work was partially supported by Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo Grants 93/3552-9 and 95/6206-0, and Conselho Nacional de Pesquisas/PADCT 62.0099/95.0.
ABBREVIATION
- GFP
green fluorescent protein
References
- 1.Mermall V, Post P L, Mooseker M S. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara M, Horiuchi H, Ohta A, Takagi M. Biochem Biophys Res Commun. 1997;236:75–78. doi: 10.1006/bbrc.1997.6907. [DOI] [PubMed] [Google Scholar]
- 3.Cheney R E, O’Shea M K, Heuseur J E, Coelho M V, Wolenski J S, Espreafico E M, Forscher P, Larson R E, Mooseker M S. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 4.Nascimento A A C, Cheney R E, Tauhata S B F, Larson R E, Mooseker M S. J Biol Chem. 1996;271:17561–17569. doi: 10.1074/jbc.271.29.17561. [DOI] [PubMed] [Google Scholar]
- 5.Mercer J A, Seperack P K, Strobel M C, Copeland N G, Jenkins N A. Nature (London) 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 6.Johnston G C, Prendergast J A, Singer R A. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haarer B K, Petzold A, Lillie S H, Brown S. J Cell Sci. 1994;107:1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- 8.Espreafico E M, Cheney R E, Matteoli M, Nascimento A A C, De Camilli P V, Larson R E, Mooseker M S. J Cell Biol. 1992;119:1541–1558. doi: 10.1083/jcb.119.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders G, Lichte B, Meyer H E, Kilimann M W. Fed Eur Biochem Soc. 1992;311:295–298. doi: 10.1016/0014-5793(92)81123-4. [DOI] [PubMed] [Google Scholar]
- 10.Zhao L-P, Koslovsky J S, Reinhard J, Bähler M, Witt A E, Provance D W, Jr, Mercer J A. Proc Natl Acad Sci USA. 1996;93:10826–10831. doi: 10.1073/pnas.93.20.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provance D W, Wei M, Ipe V, Mercer J A. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagishi Y, Oda S-I, Hayasaka S, Dekker-Ohno K, Shikata T, Inouye M, Yamamura H. Neurosci Lett. 1996;215:169–172. doi: 10.1016/0304-3940(96)12967-0. [DOI] [PubMed] [Google Scholar]
- 13.Dekker-Ohno K, Hayasaka S, Takagishi Y, Oda S-I, Wakasugi N, Mikoshiba K, Inouye M, Yamamura H. Brain Res. 1996;714:226–230. doi: 10.1016/0006-8993(95)01560-4. [DOI] [PubMed] [Google Scholar]
- 14.Coling D E, Espreafico E M, Kachar B. J Neurocytol. 1997;26:113–120. doi: 10.1023/a:1018523827852. [DOI] [PubMed] [Google Scholar]
- 15.Titus M A. Curr Biol. 1997;7:R301–R304. doi: 10.1016/s0960-9822(06)00143-6. [DOI] [PubMed] [Google Scholar]
- 16.Nascimento A A C, Amaral R G, Bizario J C S, Larson R E, Espreafico E M. Mol Biol Cell. 1997;8:1971–1988. doi: 10.1091/mbc.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Bowers B, Wei Q, Kocher B, Hammer J A., III J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- 18.Fidler I J. Nat New Biol. 1973;245:148–149. doi: 10.1038/newbio242148a0. [DOI] [PubMed] [Google Scholar]
- 19.Pawelek J, Sansone M, Koch N, Christie G, Halaban R, Hendee J, Lerner A B, Varga J M. Proc Natl Acad Sci USA. 1975;72:951–955. doi: 10.1073/pnas.72.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy D D, Segal M. J Neurosci. 1996;16:4059–4068. doi: 10.1523/JNEUROSCI.16-13-04059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolenski J S, Cheney R E, Mooseker M S, Forscher P. J Cell Sci. 1995;108:1489–1496. doi: 10.1242/jcs.108.4.1489. [DOI] [PubMed] [Google Scholar]
- 22.Baron A T, Salisbury J T. J Cell Biol. 1988;107:2669–2678. doi: 10.1083/jcb.107.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren G. Annu Rev Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- 24.Warren G. Philos Trans R Soc London B Biol Sci. 1995;349:291–295. doi: 10.1098/rstb.1995.0115. [DOI] [PubMed] [Google Scholar]
- 25.Allan V J, Vale R D. J Cell Biol. 1991;113:347–359. doi: 10.1083/jcb.113.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niclas J, Allan V J, Vale R D. J Cell Biol. 1996;133:585–593. doi: 10.1083/jcb.133.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickett-Heaps J D. Ann NY Acad Sci. 1975;253:352–361. doi: 10.1111/j.1749-6632.1975.tb19213.x. [DOI] [PubMed] [Google Scholar]
- 28.Brinkley B R. Annu Rev Cell Biol. 1985;1:145–172. doi: 10.1146/annurev.cb.01.110185.001045. [DOI] [PubMed] [Google Scholar]
- 29.Kellogg D R, Moritz M, Alberts B M. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- 30.Larson R E. Braz J Med Biol Res. 1996;29:309–318. [PubMed] [Google Scholar]
- 31.Welsh M J, Dedman J R, Brinkley B R, Means A R. Proc Natl Acad Sci USA. 1978;75:1867–1871. doi: 10.1073/pnas.75.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron A T, Greenwood T M, Salisbury J L. Cell Motil Cytoskeleton. 1991;18:1–14. doi: 10.1002/cm.970180102. [DOI] [PubMed] [Google Scholar]
- 33.Byers B. In: Molecular Genetics in Yeast. Von Wettstein D, Friis J, Keilland-Brandt M, Stenderup A, editors. Vol. 17. Copenhagen: Munksgaard; 1981. pp. 119–131. [Google Scholar]
- 34.Baum P, Furlong C, Byers B. Proc Natl Acad Sci USA. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu K P, Means A R. Endocr Rev. 1993;14:40–58. doi: 10.1210/edrv-14-1-40. [DOI] [PubMed] [Google Scholar]
- 36.Coelho M V, Larson R E. Braz J Med Biol Res. 1993;26:465–472. [PubMed] [Google Scholar]
- 37.Seperack P K, Mercer J A, Strobel M C, Copeland N G, Jenkins N A. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastural E, Barrat F J, Dufourcq-Lagelouse R, Certain S, Sanal O, Jabado N, Seger R, Griscelli C, Fischer A, De Saint Basile G. Nat Genet. 1997;16:289–292. doi: 10.1038/ng0797-289. [DOI] [PubMed] [Google Scholar]
- 39.Geiger B, Rosen D, Berke G. J Cell Biol. 1982;95:137–143. doi: 10.1083/jcb.95.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue S. Cell Struct Funct. 1996;21:375–379. doi: 10.1247/csf.21.375. [DOI] [PubMed] [Google Scholar]
- 41.Kellogg D R. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- 42.Fujiwara K, Pollard T D. J Cell Biol. 1976;71:848–875. doi: 10.1083/jcb.71.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maupin P, Pollard T. J Ultrastruct Mol Struct Res. 1986;94:92–103. doi: 10.1016/0889-1605(86)90055-8. [DOI] [PubMed] [Google Scholar]
- 44.Komesli S, Tournier F, Paintrand M, Margolis R, Job D, Bornens M. J Cell Biol. 1989;109:2869–2878. doi: 10.1083/jcb.109.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klotz C, Dabauvalle M C, Paintrand M, Weber T, Bornens M, Karsenti E. J Cell Biol. 1990;110:405–415. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley C A, Sellers J R, Gard D L, Bui D, Adelstein R S. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lillie S, Brown S. J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lillie S, Brown S. Nature (London) 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- 49.Benashski S E, Harrison A, Patel-King R S, King S M. J Biol Chem. 1997;272:20929–20935. doi: 10.1074/jbc.272.33.20929. [DOI] [PubMed] [Google Scholar]
- 50.Crepieux P, Kwon H, Leclerc N, Spencer W, Richard S, Lin R, Hiscott J. Mol Cell Biol. 1997;17:7375–7385. doi: 10.1128/mcb.17.12.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]