Abstract
Murine neonates typically mount Th2-biased immune responses. This entails a cell-intrinsic component whose molecular basis is unknown. We found that neonatal CD4+ T cells are uniquely poised for rapid Th2 function. Within 24 h of activation neonatal CD4+ cells made high levels of interleukin-4 and interleukin-13 mRNA and protein. The rapid high-level interleukin-4 production arose from a small sub-population of cells, did not require cell cycle entry, and was unaffected by pharmacologic DNA demethylation. CpG methylation analyses in resting neonatal cells revealed pre-existing hypomethylation at a key Th2 cytokine regulatory region, termed conserved noncoding sequence 1 (CNS-1). Robust Th2 function and increased CNS-1 demethylation was a stable property that persisted in neonatal Th2 effectors. The transcription factor STAT6 was not required for CNS-1 demethylation, and this state was already established in neonatal CD4 single positive thymocytes. CNS-1 demethylation levels were much greater in IL-4-expressing CD4 SP thymocytes compared to unactivated cells. Together, these results indicate that neonatal CD4+ T cells possess distinct qualities that could predispose them toward rapid, effector-like Th2 function.
INTRODUCTION
In murine neonates, T helper (Th) cell responses to protein antigens are typically Th2-dominant (1). Strong Th2 responsiveness seems to be intrinsic to neonatal cells, as made evident by experiments that have separated neonatal CD4+ T cells from the neonatal environment. For instance, unlike adult cells, neonatal T cells produce high levels of the Th2 cytokine IL-4 within 48 h of in vitro activation with adult antigen-presenting cells (APC) (2). Moreover, adoptively transferred neonatal CD4+ cells demonstrate enhanced Th2 function in vivo, when compared to transferred adult cells (3). Thus, Th2 immune deviation in neonates is increasingly understood at the cellular level, yet the molecular mechanisms responsible for this phenomenon are largely unknown.
It is striking that naïve neonatal T cells produce large amounts of IL-4 relatively early after activation, since adult cells generally require several days to weeks of development to express high levels of Th2 cytokines. This suggests that neonatal cells, unlike naïve adult cells, are pre-programmed for rapid, robust Th2 function. The acquisition of the capacity for high-level Th2 cytokine expression has been extensively studied in adult CD4+ cells. The Th2 cytokine locus contains the Il5, Il13, and Il4 genes arranged in tandem on a roughly 150 kb region of mouse chromosome 11. In naïve adult CD4+ cells, this locus exists in a transcriptionally silent state characterized by condensed chromatin and locus-wide hypermethylation of CpG residues. Activation under Th2-polarizing conditions leads to subsequent remodeling of the locus and high-level transcription of the Th2 cytokine genes (4). The transition of the Th2 cytokine locus to a fully-open, highly-transcribed state involves several processes including cellular proliferation (5, 6), the upregulation of lineage-determining transcriptions factors (7), the appearance of DNase I hypersensitivity sites (8-11), histone modifications (11-15), and extensive DNA demethylation (10, 11, 14, 16, 17). While the various processes controlling Th2 differentiation and function have been extensively studied in adult cells, it is not known how naïve neonatal CD4+ cells achieve such high levels of Th2 function early after activation.
There is ample evidence that DNA methylation is an important regulator of Th2 cytokine expression in adult CD4+ cells. Several laboratories have demonstrated that the murine Th2 cytokine locus becomes demethylated as naïve adult CD4+ cells differentiate into effector cells (6, 11, 14, 16, 17). Demethylation proceeds with different kinetics at the various parts of the locus (6, 11, 14, 16, 17). Some regulatory regions, such as rad50 RHS7 within the newly identified locus control region (LCR), become almost fully demethylated within several days of Th2 polarization (11). Other regulatory regions, such as CNS-1, are only partially demethylated within the same time frame (16). In contrast, non-regulatory regions within the Il4 gene require weeks of Th2 polarization before complete demethylation is achieved (16). The clear importance of DNA methylation as a non-redundant repressor of Th2 cytokine expression has recently been shown in adult mice deficient for the maintenance DNA methyltransferase Dnmt1. In these mice, Il4, Il13, and Il5 expression are markedly enhanced (18).
In this report, we focused on the prototypical Th2 cytokines IL-4 and IL-13 in order to understand the molecular regulation of Th2 cytokine expression in neonatal CD4+ T cells. We found that recently activated neonatal CD4+ cells expressed higher levels of Il4 and Il13 mRNA and protein compared to adult cells. Using the bisulfite genomic sequencing technique (19), we discovered that a coordinate regulatory region of Th2 cytokine expression, CNS-1 (20), was uniquely demethylated in resting neonatal CD4+ cells. The transcription factor STAT6 was dispensable for CNS-1 demethylation, and this state was already established in neonatal CD4 single positive (SP) thymocytes. IL-4-expressing neonatal thymocytes exhibited substantially greater levels of CNS-1 demethylation compared to unactivated cells. Our data suggests that the relatively hypomethylated state of CNS-1 in neonatal CD4+ cells facilitates their rapid, high-level expression of the Il4 and Il13 genes. Thus, the developmental regulation of epigenetic patterns at the Th2 cytokine locus may serve to maintain an anti-inflammatory state in early life.
METHODS
Mice
Wild-type, STAT6-deficient, and DO11.10 TCR-transgenic mice on a BALB/c background were bred and housed in pathogen-free conditions in the Division of Veterinary Resources at the University of Miami Miller School of Medicine. All animal studies were approved by the University of Miami Animal Care and Use Committee. Neonatal animals were 7 days old and adult mice were 6-10 weeks old. For the CD4 SP thymocyte experiments, neonates were 2 days old.
Cell culture
Cell suspensions were made from pools of mesenteric, inguinal, axillary, brachial, and cervical lymph nodes. CD4+ cells were positively selected from the cell suspensions on Miltenyi Biotec (Auburn, CA) MACS MS+ columns using the manufacturer’s protocol. Cell purity was routinely >97%. For the DNA methylation studies, neonatal and adult CD4+ cells were stained with anti-CD4 CyChrome (BD Biosciences Pharmingen, San Diego, CA) and anti-CD44 biotin (Caltag Laboratories, Burlingame, CA), followed by PE-coupled streptavidin (Jackson Immunoresearch, West Grove, PA). CD4+CD44low cells were purified on a Vantage SE cell sorter (BD Biosciences, Mountain View, CA). For the neonatal thymocyte experiments, the cells were stained with anti-CD4 FITC (Caltag) and anti-CD8 PE (Pharmingen), followed by sorting for CD4 SP thymocytes.
2 × 105 CD4+ cells were activated in 96 well plates with 10 μg/ml pre-coated anti-CD3ε (mAb 1452C11, purified from ascites) and 0.5 μg/ml soluble anti-CD28 (mAb 37.51, Pharmingen). To measure antigen specific responses, 2 × 105 CD4+ cells from DO11.10 mice were activated in the presence of 4 × 105 adult splenic APC (APC preparation previously described in (2)) and various concentrations of chicken OVA peptide (A.A. 323-339). Culture medium contained RPMI 1640 (Life Technologies, Grand Island, NY) supplemented with 1 mM sodium pyruvate (Life Technologies), 2 mM l-glutamine (Life Technologies), 5 × 10-2 mM 2-ME (Life Technologies), 1% penicillin-streptomycin (Life Technologies), and 10% heat-inactivated (56°C, 30 min.) FCS (HyClone, Logan, UT).
To generate Th2 effectors, activated CD4+ cells were cultured with 10 ng/ml rIL-2 (R&D Systems, Minneapolis, MN), 25 ng/ml rIL-4 (R&D Systems), and 10 μg/ml neutralizing anti-IFN-γ mAb (XMG1.2, purified from ascites). Th1 effectors were generated with 10 ng/ml rIL-2, 10 ng/ml rIL-12 (Peprotech Inc., Rocky Hill, NJ), and 1 μg/ml neutralizing anti-IL-4 mAb (11B11, purified from ascites). After 3 days, the cells were washed and then expanded in rIL-2 for 2 more days.
Cell cycle arrest was achieved pharmacologically using 5 μg/ml aphidicolin (Sigma-Aldrich, St. Louis, MO), 500 μg/ml hydroxyurea (Sigma-Aldrich), or 200 ng/ml paclitaxel (Sigma-Aldrich). For pharmacologic DNA demethylation, 1 μM 5-aza-deoxycytidine (Sigma-Aldrich) was added at the beginning of cell culture.
Cytokine ELISPOT and ELISA
After 20 h or 5 days in culture, activated CD4+ cells were harvested, washed, and suspended in medium containing 10 nM PMA (Sigma-Aldrich) and 0.25 μM ionomycin (Calbiochem, La Jolla, CA). The cells were then cultured for 4 h in Nunc Maxisorp plates (Nunc, Naperville, IL) pre-coated with the anti-IL-4 mAb 11B11 (Pharmingen) and the plates were processed for ELISPOT analyses, as previously described (21).
The IL-4 (Pierce-Endogen, Rockford, IL), IL-13 (R&D Systems), or IL-2 (Pierce-Endogen) content of culture supernatants was measured using specific cytokine ELISA kits as described elsewhere (2, 22).
Cell staining
For the cell cycle arrest or 5-aza-deoxycytidine experiments, resting and activated cells were stained with anti-CD69 biotin (Pharmingen) or anti-CD25 biotin (Caltag Laboratories), respectively, followed by FITC-coupled streptavidin (Pharmingen). The cells were then processed for propidium iodide (Sigma-Aldrich) staining as previously described (23).
For the intracellular cytokine staining experiments, activated CD4+ cells were washed and cultured for 4 h in medium containing 10 nm PMA, 0.25 μM ionomycin, and 1 μM monensin (Ebioscience, San Diego, CA). The cells were then washed and fixed for 20 min in 250 μl Cytofix/Cytoperm (Pharmingen). Resting cells were fixed concurrently with 250 μl Cytofix/Cytoperm. After two washes with 1× Perm/Wash buffer (Pharmingen), the cells were stained with anti-IL-4 PE (mAb 11B11, Pharmingen) and anti-IFN-γ FITC (mAb XMG1.2, Pharmingen) for 30 min. Isotype control staining for IFN-γ was performed in parallel using a FITC-coupled rat IgG1 mAb A110-1 (Pharmingen). The specificity of IL-4 staining was determined by incubating the cells with an unlabeled anti-IL-4 mAb 11B11 (purified from ascites) for 30 min, followed by a PE-coupled anti-IL-4 mAb 11B11 for another 30 min. After staining, the cells were washed twice with 1× Perm/Wash buffer followed by a single wash with staining buffer.
RNA isolation and RT-PCR
Total RNA was isolated using RNeasy Mini Kits (Qiagen, Valencia, CA). 5 μg of total RNA was reverse-transcribed in reactions containing 125 U of Powerscript reverse transcriptase (BD Biosciences Clontech, Palo Alto, CA), 1× First Strand buffer (Clontech), 10 U recombinant RNase inhibitor (Clontech), 1 mM Ultrapure dNTP mix (Clontech), and 3.8 μM Oligo dT16 primer (Applied Biosystems, Foster City, CA). The conditions for the reaction were as follows: 10 min at 70°C, 90 min at 42°C, and 15 min at 70°C. For the quantitative RT-PCR assays, cDNA from reactions containing 125-250 ng of input total RNA was amplified in PCR reactions using LightCycler® Fast DNA SYBR Green I kits (Roche Diagnostics, Indianapolis, IN) and a Roche LightCycler® in accordance with the manufacturer’s instructions. Primer sequences for Il4, Il13, and Actb are listed below. The cycling parameters for both Il4 and Actb were as follows: 1 cycle at 95°C for 10 min; 36 cycles at 95°C for 10 s, 65°C for 8 s, 72°C for 20 s, and a single acquisition point for each cycle after 10 s at 85°C. For Il13, the cycling parameters were as follows: 1 cycle at 95°C for 10 min; 33 cycles at 95°C for 10 s, 57°C for 20 s, 72°C for 18 s, and a single acquisition point for each cycle after 10 s at 85°C. Standard curves for Il4, Il13, and Actb were generated using serial dilutions of plasmid DNA containing cloned PCR products for each gene. Using this method, the amplification efficiencies were determined to be equivalent for the Il4, Il13, and Actb primers. For each sample, the absolute levels of Il4, Il13, and Actb were extrapolated from the standard curves using Roche LightCycler® data analysis software version 3.5. The Il4 or Il13 levels were normalized to those of Actb for individual samples. The specificity of the reactions was determined by melting curve analysis and by agarose gel electrophoresis. Control reactions lacking reverse transcriptase ensured that the products were derived from RNA templates. All reactions were performed in triplicate at the UM/Sylvester Molecular Core Analysis Facility at the University of Miami Miller School of Medicine.
DNA isolation and bisulfite sequencing
Genomic DNA was isolated using QIAamp DNA Mini Kits (Qiagen). The DNA was sheared by passage through a 30-gauge needle 20 times followed by denaturation in 0.3 M NaOH for 20 min at 50°C. Sodium bisulfite modification was performed with CpGenome Kits (Chemicon International, Temecula, CA) using the manufacturer’s protocol. Bisulfite modified DNA was amplified via touchdown PCR in reactions containing 0.625 U of iTaq DNA Polymerase (Biorad), 1× PCR buffer (Biorad), 1.5 mM MgCl2 (Biorad), 0.2 mM Ultrapure dNTP mix, and 0.25 μM primers specific for Il4 intron I-exon 2, the CNS-1 region, the consensus sequence 1 (CS-1) region of the Il13 promoter, or RHS7 of the rad50 gene. The cycling parameters were as follows:
Il4intron I-exon 2: 1 cycle at 95°C for 5 min; 5 touchdown cycles decreasing the annealing temperature 2°C per cycle at 95°C for 30 s, 70-64°C for 30 s, and 72°C for 45 s; 35 cycles at 95°C for 30 s, 62°C for 30 s, and 72°C for 45 s; 1 cycle at 72°C for 7 min.
CNS-1: 1 cycle at 95°C for 5 min; 5 touchdown cycles decreasing the annealing temperature 1°C per cycle at 95°C for 30 s, 60-56°C for 30 s, and 72°C for 45 s; 40 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s; 1 cycle at 72°C for 7 min.
Il13CS-1: 1 cycle at 95°C for 5 min; 5 touchdown cycles decreasing the annealing temperature 1°C per cycle at 95°C for 30 s, 63-59°C for 30 s, and 72°C for 45 s; 35 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 45 s; 1 cycle at 72°C for 7 min.
rad50RHS7: 1 cycle at 95°C for 5 min; 5 touchdown cycles decreasing the annealing temperature 2°C per cycle at 95°C for 30 s, 62-54°C for 30 s, and 72°C for 45 s; 40 cycles at 95°C for 30 s, 53°C for 30 s, and 72°C for 45 s; 1 cycle at 72°C for 7 min.
Primer sequences
The nucleotide sequences for Il4 Forward and Reverse RT-PCR primers (24), Il13 Forward and Reverse RT-PCR primers (25), and CNS-1 oligonucleotides #1 and #2 (26) used for bisulfite sequencing were derived from the literature as cited.
A. Real-time RT-PCR
Il4Forward: 5’-ACGGCACAGAGCTATTGATG-3’ Il4Reverse: 5’-TACCACCGAGTCATGATGCT-3’
ActbForward: 5’-GTGGGCCGCTCTAGGCACCAA-3’ ActbReverse: 5’-GAGAAACTACAGTGCGTGCTAAAG-3’
Il13Forward: 5’-CTTGCTTGCCTTGGTGGTCTCGC-3’ Il13Reverse: 5’-AGCCCACTTTATAACAAAACTGC-3’
B. Bisulfite Sequencing
Il4intron I-exon 2 oligo #1: 5’-TGGTTGAGGTAGGATTAGGGATAAAG-3’ Il4intron I-exon 2 oligo #2: 5’-CCTCCAAAATATACCACAACAAACC-3’
CNS-1 oligo #1: 5’-TATAAGGGTTGTAGGAAGAGTAATGTAGTTTTTATAT-3’ CNS-1 oligo #2: 5’-CACTTTAAACTAATCAAACCTAAAACTATATCCATA-3’
Il13promoter CS-1 oligo #1: 5’-GGTAAGGTGGTAGATTTTTTGGAG-3’ Il13promoter CS-1 oligo #2: 5’-AAATTCTCAATTCTCCCACTACAAC-3’
rad50RHS7 oligo #1: 5’-AAGTTGTTATTGATGTAGTAGAAAG-3’ rad50RHS7 oligo #2: 5’-CTTACCTCTCCCCAAATATAC-3’
Sorting of IL-4-expressing cells
CD4 SP thymocytes from day 3 neonatal DO11.10 mice were enriched by negative selection using Miltenyi CD8 beads followed by passage through Miltenyi MACS MS+ columns. 4 × 105 cells were activated with 10 μg/ml plate-bound anti-CD3ε mAb and 0.5 μg/ml soluble anti-CD28 mAb in the presence of 10 ng/ml rIL-2. After 2 days, the cells were harvested, washed, and suspended in media solution containing 10 nM PMA and 0.25 μM ionomycin. After 2 h, the thymocytes were stained for IL-4 expression using Miltenyi IL-4 Cell Enrichment and Detection Kits in accordance with the manufacturer’s instructions. Following additional staining with anti-CD4 mAb, CD4+IL-4+ cells were sorted on a BD Biosciences FACSARIA. The cells were then prepared for lysis and DNA isolation as described in detail above. Bisulfite genomic DNA conversion was performed as previously described by Chris Wilson’s laboratory (26).
RESULTS
Neonatal CD4+ cells produce high levels of IL-4 and IL-13 early after activation
We have previously observed that freshly-activated naïve total T cells from neonatal mice produce much greater levels of IL-4 than do adult cells when stimulated with soluble anti-CD3ε and adult APC (2). These findings suggest that enhanced Th2 function is an intrinsic property of neonatal T cells. In order to study this at the molecular level, we first needed to examine Th2 cytokine expression in purified neonatal CD4+ T cells. Thus, we activated DO11.10 TCR-transgenic neonatal or adult CD4+ cells with plate-bound anti-CD3ε and soluble anti-CD28 mAb. After 24 h, the levels of IL-4 or IL-13 protein in the culture supernatants were measured by ELISA. Compared to adult cells, neonatal CD4+ cells produced greater levels of both IL-4 (4.4-16.0 fold higher) and IL-13 (1.2-24.9 fold higher) at this early timepoint (Fig. 1a).
Figure 1. Murine neonatal CD4+ cells produce high levels of IL-4 and IL-13 early after activation.

CD4+ cells were isolated from day 7 neonatal or adult TCR-transgenic DO11.10 mice. The cells were activated with plate-bound anti-CD3ε plus soluble anti-CD28 mAb (polyclonally). (a) After 24 h, the IL-4 (left graph) or IL-13 (right graph) content in culture supernatants was measured by ELISA. The fold difference in IL-4 or IL-13 secretion between neonatal and adult cells is indicated for each experiment. (b) After 24 h, total RNA was isolated from activated neonatal or adult CD4+ cells. The RNA was reverse transcribed and cDNA products were quantified via real-time PCR using SYBR Green I detection reagents and primers specific for Il4, Il13, or Actb. The levels of Il4 or Il13 mRNA were normalized to those of Actb before making comparisons between samples. The sample with the lowest expression of Il4 (left graph) or Il13 (right graph) was assigned an expression value of 1. The expression values of the other samples are relative to those of the lowest sample. Each reaction was performed in triplicate for both of the depicted experiments. (c) Total RNA was prepared from freshly-isolated neonatal DO11.10 CD4+ cells or from neonatal DO11.10 CD4+ cells that were activated with plate-bound anti-CD3ε plus soluble anti-CD28 mAb for 24 h. Quantitative RT-PCR for Il4 (left graph) and Il13 (right graph) was performed as described above. (d) CD4+ lymph node cells from day 7 neonatal or adult TCR-transgenic DO11.10 mice were activated with cognate ova peptide. After 48 hr, the IL-4 (left graph) or IL-2 (right graph) content in culture supernatants was measured by ELISA. The fold difference in IL-4 or IL-2 secretion between neonatal and adult cells is indicated for each experiment. All results in (a), (b), (c), and (d) are graphed as the mean ± the standard deviation of the triplicate values.
We next tested whether the differences in IL-4 and IL-13 protein production between activated neonatal and adult CD4+ cells were also evident at the mRNA level. Quantitative RT-PCR analysis revealed more abundant Il4 (4.2-18.5 fold higher) and Il13 (3.7-5.1 fold higher) transcripts in neonatal cells, compared to adult cells, after 24 h of activation (Fig. 1b). The differences in Il4 or Il13 mRNA between activated neonatal and adult cells were within the same range as the differences in IL-4 and IL-13 protein measured in culture supernatants by ELISA (Fig. 1a). Moreover, activation of neonatal cells resulted in a de novo increase in Il4 and Il13 mRNA (Fig. 1c), suggesting that Th2 cytokine expression is transcriptionally regulated in neonatal CD4+ T cells, as it is in adult cells (27).
To determine whether antigen-specific Th2 responses were also enhanced in neonatal cells, neonatal and adult CD4+ cells from DO11.10 TCR-transgenic mice were activated with chicken ova peptide in the presence of adult splenic APC. Two days later, ova-specific cytokine responses were measured by ELISA. Neonatal CD4+ cells clearly made more IL-4 (range 2.7-16.3 fold higher levels) than did adult cells at all concentrations of ova peptide tested (Fig. 1d). In contrast, neonatal and adult cells made similar levels of IL-2. These findings indicate that recently-activated, naïve neonatal CD4+ cells mount greater antigen-specific IL-4 responses, but similar IL-2 responses, compared to adult cells.
Greater frequencies of neonatal than adult CD4+ cells produce IL-4 early after activation
The differences in IL-4 production between recently activated neonatal and adult CD4+ cells could potentially be explained by differences in the frequencies of responding cells. To investigate this possibility, intracellular cytokine staining was performed on neonatal or adult DO11.10 CD4+ cells after 24 h of activation (Fig. 2a). While the frequencies of neonatal and adult IFN-γ-producing cells were similar, only neonatal cells stained positively for IL-4. Despite their early high-level Th2 function (Fig. 1a), the frequencies of IL-4-producing neonatal cells were surprisingly low, ranging from 0.5-1.1% at this timepoint (Fig. 2a and data not shown). IL-4 and IFN-γ were not detectable in freshly-isolated neonatal CD4+ cells, suggesting that neonatal cells do not contain pre-formed stores of intracellular IL-4 protein, as has previously been described for mast cells (28).
Figure 2. Greater frequencies of neonatal than adult CD4+ cells produce IL-4 early after activation.

CD4+ cells were isolated from day 7 neonatal or adult DO11.10 mice. (a) Intracellular staining for IL-4 and IFN-γ was performed on freshly-isolated resting cells (left column) or on cells that were polyclonally activated for 20 h (right column), followed by 4 h of culture with PMA, ionomycin, and monensin. To ensure the specificity of the staining, identically treated cells were stained with control antibodies (smaller inlaid graphs). The depicted experiment is representative of 3 separate experiments. (b) Neonatal or adult CD4+ cells were polyclonally activated for 20 h, washed, and transferred to ELISPOT wells containing PMA and ionomycin for 4 additional h. The frequencies of IL-4-secreting cells were enumerated by ELISPOT. Each sample was plated in quadruplicate and the averages ± the standard deviations are shown. The fold difference in the frequencies of IL-4-producing neonatal and adult CD4+ cells is indicated for each experiment.
Since IL-4 was undetectable in activated adult cells by intracellular staining, we used a more sensitive assay, the ELISPOT, to compare differences in the frequencies of IL-4-secreting neonatal and adult CD4+ T cells (Fig. 2b). After 24 h of activation, the frequencies of IL-4-secretors were 4.6-12.2 fold higher among neonatal CD4+ cells compared to adult cells. This range is similar to the relative differences in total IL-4 protein or Il4 mRNA at 24 h (Fig. 1). The frequencies of IL-4-producing neonatal CD4+ cells ranged from roughly 0.5-2% at this timepoint (Fig. 2b), corroborating our intracellular cytokine staining results (Fig. 2a). Therefore, the robust early IL-4 responses of murine neonates arise from a small population of IL-4-secreting CD4+ cells that are nonetheless present in greater numbers in comparison with adult mice.
Early IL-4 production by neonatal CD4+ cells does not require cell cycle entry
It has previously been shown that IL-4 expression increases with successive cell divisions as naïve adult CD4+ cells develop into Th2 effectors (5, 6). This is thought to coincide with the opening of the Th2 cytokine locus and the stable imprinting of epigenetic modifications that are propagated to daughter cells with each division. Once these modifications are established, adult Th2 effectors rapidly express IL-4 in a cell cycle-independent manner (6). Thus far, our findings have indicated that at least a subset of neonatal CD4+ cells exist in a state that is permissive for high-level IL-4 production early after activation. Therefore, it is possible that neonatal cells resemble adult Th2 effectors by making IL-4 without entering the cell cycle. To test this hypothesis, neonatal DO11.10 CD4+ cells were activated in the presence of the cell cycle inhibitors aphidicolin (G1/S arrest), hydroxyurea (early S arrest), or paclitaxel (G2/M arrest) and the IL-4 content of the culture supernatants was measured by ELISA after 24 h. When treated with these agents, neonatal cells produced levels of IL-4 that were slightly lower than those made by untreated controls (Fig. 3a). This IL-4 production was not due to incomplete arrest, since the inhibitors blocked neonatal cells at the appropriate stages of the cell cycle (Fig. 3b). Moreover, the treated cells were activated to a similar degree as untreated controls, as evidenced by their equivalent expression of the early activation marker CD69 (Fig. 3c). Taken together, these findings indicate that early IL-4 production by neonatal cells does not require cell cycle entry.
Figure 3. Early IL-4 production by neonatal CD4+cells does not require cell cycle entry.

CD4+ cells from day 7 neonatal DO11.10 mice were activated polyclonally for 24 h. Some cultures contained the cell cycle inhibitors aphidicolin (G1/S arrest), hydroxyurea (early S arrest), or paclitaxel (G2/M arrest). (a) The IL-4 content in culture supernatants was determined by ELISA. The results are graphed as the mean ± the standard deviation of the triplicate values from a representative experiment. (b) After ethanol fixation, the cells were stained with propidium iodide to measure the percentages of apoptotic cells (left marker) and actively cycling cells (S + G2/M, right marker). (c) Prior to propidium iodide staining, the cells were stained for surface expression of the early activation marker CD69 (solid line). Unactivated cells were stained as controls (dotted line). The experiment in (c) was electronically gated on non-apoptotic cells. All results are representative of at least 3 independent experiments.
Pharmacologic demethylation does not affect IL-4 production by neonatal CD4+ cells
DNA methylation is an epigenetic silencing mechanism whose manner of inheritance is inexorably linked to the cell cycle. After associating with replicating loci during S phase, the maintenance DNA methyltransferase Dnmt1 faithfully copies DNA methylation patterns from the parental to the newly-synthesized DNA strand (29). It is intriguing that neonatal CD4+ cells still made moderately high levels of IL-4 despite inhibition of DNA synthesis with the DNA polymerase inhibitor aphidicolin (Fig. 3a). This raised the possibility that neonatal cells could already be hypomethylated at the Th2 cytokine locus, and thus would be unaffected by the repressive effects of maintenance DNA methylation. To test this hypothesis, neonatal or adult CD4+ cells from DO11.10 mice were activated in the presence of the demethylating agent 5-aza-2’-deoxycytidine (5-aza-deoxycytidine) and the IL-4 content of the culture supernatants was measured by ELISA after 72 h. As expected (6, 14), IL-4 production was greatly increased in treated adult cells relative to untreated controls (Fig. 4a). In contrast, IL-4 production by neonatal cells was unchanged in treated cells compared to untreated controls (Fig. 4a). The differences between neonatal and adult cells were specific for IL-4 since production of the Th1 cytokine IFN-γ was enhanced by 5-aza-deoxycytidine treatment to a similar degree in neonatal and adult cells (Fig. 4b).
Figure 4. Pharmacologic demethylation does not affect IL-4 production by neonatal CD4+ cells.

CD4+ cells from day 7 neonatal or adult DO11.10 mice were activated polyclonally in the presence or absence of the demethylating agent 5-aza-deoxycytidine. After 72 h, the (a) IL-4 or (b) IFN-γ content in culture supernatants was determined by ELISA. The results are graphed as the mean ± the standard deviation of the triplicate values from a representative experiment. The fold difference in (a) IL-4 or (b) IFN-γ secretion between treated and untreated neonatal or adult cells is indicated. (c) After 48 h, 5-aza-deoxycytidine (5-aza-dc) treated cells (filled histogram) or untreated cells (open histogram) were stained for surface expression of the activation marker CD25. Unactivated cells were also stained as controls (dotted line). (d) After 48 h, treated and untreated cells were fixed and stained with propidium iodide. The percentages of apoptotic cells (left marker) and actively cycling cells (S + G2/M, right marker) are indicated. The experiment in (c) was electronically gated on non-apoptotic cells. All results are representative of 4 independent experiments.
Differences in cell activation, apoptosis, or cycling could trivially account for the distinct IL-4 responses of neonatal and adult cells to treatment with this agent. Therefore, we investigated the cellular activation status and the percentages of apoptotic cells and cells in cycle after 48 or 72 h of culture with 5-aza-deoxycytidine. For both neonatal and adult cells, expression of the activation marker CD25 was comparable between treated and untreated cells (Fig. 4c and data not shown). Higher percentages of both neonatal and adult cells underwent apoptosis when treated with 5-aza-deoxycytidine relative to their respective untreated controls (Fig. 4d). However, the percentages of apoptotic cells were similar between treated neonatal and adult cells at both 48 and 72 h (Fig. 4d and data not shown). The percentages of cells in the S, G2, and M phases of the cell cycle were also comparable between neonatal and adult cells whether or not they were treated with 5-aza-deoxycytidine (Fig. 4d and data not shown). Altogether, these findings indicate that the demethylating effects of 5-aza-deoxycytidine did not affect IL-4 production by neonatal CD4+ cells, suggesting that neonatal cells are less methylated at the Th2 cytokine locus than are adult cells.
The CNS-1 region pre-exists in a demethylated state in resting neonatal CD4+ cells
Pharmacologic demethylation of neonatal and adult CD4+ T cells (Fig. 4a) suggested that neonatal cells may be relatively hypomethylated at the Th2 cytokine locus (Fig. 5a). To test this directly, we used the bisulfite genomic sequencing method (19) to compare DNA methylation patterns between neonatal and adult 5-day DO11.10 Th2 effectors (Figs. 5b and 5c). 14 CpG sites were analyzed within a highly conserved, coordinate regulatory region of Th2 cytokine expression, termed CNS-1 (20). As expected (14, 16), CpG demethylation was detected in adult Th2 effectors (Fig. 5b). However, a comparison of DNA methylation levels at each individual CpG site revealed that demethylation was 1.5-5.6 fold greater in neonatal Th2 cells relative to adult Th2 cells (Fig. 5c). Strikingly, all 14 CpG sites were completely demethylated in 58% of the clones from neonatal Th2 effectors compared with only 7% of the clones from adult Th2 effectors (Table 1). In contrast to the Th2 effectors, adult Th1 cells exhibited little demethylation at each of the CpG sites within the CNS-1 region and none of the clones from Th1 effectors was completely demethylated (Fig. 5b). The levels of demethylation strongly reflected the capacity to express IL-4 since neonatal Th2 cells had greater numbers of IL-4-secreting cells than did adult Th2 cells and very few adult Th1 cells made IL-4 (Fig. 5d).
Figure 5. CNS-1 is highly demethylated in neonatal Th2 effectors.
(a) A schematic of the Th2 cytokine genomic locus in adult Th2 effector cells. (b) CD4+ cells were enriched from day 7 neonatal or adult DO11.10 mice. Genomic DNA was isolated from neonatal or adult 5-day Th2 effectors or adult 5-day Th1 effectors. DNA methylation at 14 CpG sites within the CNS-1 region was assessed via bisulfite sequencing. CpG sites are numbered based on sequence AC005742. (c) The percentages of demethylated clones at each CpG site and the numbers of clones sequenced in total (indicated in parentheses) are represented as pooled data from two independent cell preparations. (d) A subset of neonatal and adult effector cells was washed and cultured with PMA, ionomycin, and monensin for 4 h. The cells were stained for intracellular IL-4 and IFN-γ as in Figure 2. The depicted experiment is representative of 5 independent experiments.
Table 1.
The percentages of clones that were completely demethylated at every CpG site for each genomic region analyzed within the Th2 cytokine locus.a
| Cell Type | CNS-1 | Il4 intron I - exon 2 | rad50 RHS7 LCR | Il13 CS-1 |
|---|---|---|---|---|
| Neo Resting CD4+ | 8% | 0% | 0% | 90% |
| Adult Resting CD4+ | 0% | 0% | 0% | 95% |
| Neo Th2 | 58% | 0% | not done | not done |
| Adult Th2 | 7% | 0% | not done | not done |
CD4+ lymph node cells were enriched from day 7 neonatal or adult DO11.10 mice. Genomic DNA was isolated from naïve resting cells or from 5-day Th2 effectors. CpG methylation was assessed via the bisulfite genomic sequencing method. Four regions of the Th2 cytokine locus were analyzed: 14 CpG sites within the CNS-1 region, 6 CpG sites within intron I-exon 2 of the Il4 gene, 9 CpG sites within or adjacent to rad50 RHS7 of the Th2 cytokine LCR, and 13 CpG sites within CS-1 of the distal Il13 promoter. The percentages of DNA clones that were completely demethylated at every CpG site for each of the regions analyzed are indicated. The presented data is pooled from two independent cell preparations for each cell type.
Greater demethylation at CNS-1 was observed in neonatal CD4+ cells, relative to adult cells, after 5 days of Th2 polarization in vitro (Figs. 5b, 5c, and Table 1). This raised the intriguing possibility that resting neonatal cells could pre-exist in a demethylated state at this region. To investigate this idea, CpG methylation patterns at CNS-1 were analyzed in resting naïve adult (sorted CD4+CD44lo) and neonatal CD4+ cells from DO11.10 mice (Figs. 6a and 6b). While demethylated CpG dinucleotides were infrequent in resting adult CD4+ cells (range = 0-12% demethylated), resting neonatal cells were demethylated to varying degrees (range = 11-47% demethylated) at each of the CpG sites analyzed (Fig. 6a). Eight percent of the clones from resting neonatal CD4+ cells were demethylated at all 14 CpG sites whereas none of the clones from resting adult cells was completely demethylated (Table 1). The differences between neonates and adults were not due to the selective analysis of naïve cells in adults since similar levels of CNS-1 demethylation were observed when CD4+CD44lo naïve phenotype neonatal cells were purified from the total cell population (Fig. 6c). Thus, subsequent experiments were performed on the total resting CD4+ cell population in neonates. Taken together, these findings demonstrate that a subset of resting neonatal CD4+ cells have pre-existing demethylation at CNS-1, which could allow for high-level IL-4 and IL-13 production early after activation.
Figure 6. CpG demethylation at CNS-1 pre-exists in resting neonatal CD4+cells.

(a) As described in Figure 5, CNS-1 CpG methylation levels were assessed in naïve resting adult (sorted CD4+CD44lo) or day 7 neonatal DO11.10 CD4+ cells. (b) The percentages of demethylated clones at each CpG site and the numbers of clones sequenced in total (indicated in parentheses) are represented as pooled data from two independent cell preparations. (c) As described above, CpG methylation at CNS-1 was assessed in sorted (CD44lo) or unsorted resting CD4+ cells from day 7 DO11.10 neonates.
CpG methylation patterns at other control regions of the Th2 cytokine locus are similar in resting neonatal and adult CD4+ cells
CNS-1 was relatively demethylated in resting neonatal CD4+ cells (Fig. 6 and Table 1). However, widespread hypomethylation of the Th2 cytokine locus was not a feature of neonatal cells, since 6 CpG sites within Il4 intron I and exon 2 were only 0-18% demethylated, even in 5-day neonatal Th2 effectors (Fig. 7a and Table 1). We also tested whether neonatal cells were demethylated at another coordinate control region of Il4 and Il13 expression, rad50 RHS7 of the Th2 cytokine LCR (10, 11). As expected (11), resting naïve adult cells had minimal demethylation at 9 CpG dinucleotides within and adjacent to this region (Fig. 7b). In contrast to our findings at CNS-1 (Fig. 6), resting neonatal CD4+ cells exhibited little demethylation at rad50 RHS7 (Fig. 7b). Furthermore, none of the clones from resting neonatal or adult cells was demethylated at every CpG site (Table 1). Thus, resting neonatal CD4+ cells do not have pre-existing demethylation at rad50 RHS7 of the Th2 cytokine LCR.
Figure 7. Other regulatory regions of the Th2 cytokine locus are not differentially methylated in neonatal and adult cells.

As described in Figure 5, bisulfite methylation analysis was conducted on genomic DNA from (a), (b), (c) naïve resting cells or (a) 5-day Th2 effectors at several CpG sites within the Th2 locus including (a) 6 CpG sites within intron I and exon 2 of the Il4 gene, (b) 9 CpG sites within or adjacent to rad50 RHS7 of the Th2 cytokine LCR, or (c) 13 CpG sites within the CS-1 region of the Il13 promoter. Each CpG site is numbered based on sequence AC005742. The percentages of demethylated clones at each CpG site and the numbers of clones sequenced in total (indicated in parentheses) are represented as pooled data from two independent cell preparations.
DNA methylation levels were also analyzed at a highly conserved regulatory region containing a CpG island within the distal Il13 promoter, termed consensus sequence 1 (CS-1) (30). Since this region is an enhancer of both Il4 and Il13 promoter-driven reporter gene expression (15), it is possible that pre-existing differences in DNA methylation levels at CS-1 could contribute to the differences in Il4 and Il13 expression between recently activated neonatal and adult CD4+ cells (Figs. 1a and 1b). Individual CpG dinucleotides at this region were 95-100% demethylated in both neonatal and adult resting cells (Fig. 7c). Near complete demethylation of CpG sites extended roughly 200 bp upstream and downstream of CS-1 for both neonatal and adult cells (data not shown). In fact, at least 90% of resting neonatal or adult CD4+ cells were demethylated at every CpG site within CS-1 (Table 1). These findings demonstrate that CS-1 of the distal Il13 promoter is already demethylated in both neonatal and adult resting CD4+ cells. Therefore, greater early Il4 and Il13 expression by neonatal cells cannot be explained by pre-existing differences in CpG methylation at CS-1 in neonatal and adult cells.
The transcription factor STAT6 is not required for CNS-1 demethylation and early IL-4 expression in neonatal CD4+ cells
STAT6 is a key positive regulator of Th2 effector cell development in adult CD4+ T cells (31-33). STAT6 has been shown to be required for demethylation of a Th2 cytokine regulatory region (9). However, its role in CNS-1 demethylation and, in particular, its function in neonatal CD4+ cells is currently unknown. Thus far, our DNA methylation analyses have been performed entirely on DO11.10 TCR-transgenic CD4+ cells. Therefore, before investigating the STAT6-dependence of CNS-1 demethylation, it was first necessary to determine whether this state also occurs in resting non-transgenic neonatal cells. For this reason, CNS-1 DNA methylation patterns were compared in resting CD4+ lymph node cells from day 7 neonatal DO11.10, wild-type BALB/c, and STAT6-deficient mice on a BALB/c background. CNS-1 demethylation clearly occurred in wild-type BALB/c neonatal cells, often at levels comparable to those in DO11.10 TCR-transgenic neonatal cells (Fig. 8a). For the majority of the CpG sites within CNS-1, STAT6-deficient neonatal CD4+ cells had similar levels of DNA demethylation relative to their wild-type BALB/c counterparts (Fig. 8a). These findings indicate that CNS-1 demethylation occurs in non-transgenic neonatal CD4+ T cells and that STAT6 is not required to establish this state. To determine whether recently activated STAT6-deficient neonatal CD4+ cells are competent to make IL-4, the frequencies of IL-4-producing cells were measured by ELISPOT after 24 h of polyclonal stimulation. As in adult cells, IL-4-secreting neonatal cells were only slightly diminished among STAT6-deficient cells compared to wild-type BALB/c cells (Fig. 8b). This confirmed that, like CNS-1 demethylation, IL-4 production by recently activated neonatal CD4+ cells does not require STAT6.
Figure 8. The transcription factor STAT6 is not required for CNS-1 demethylation and early IL-4 expression in neonatal CD4+ cells.
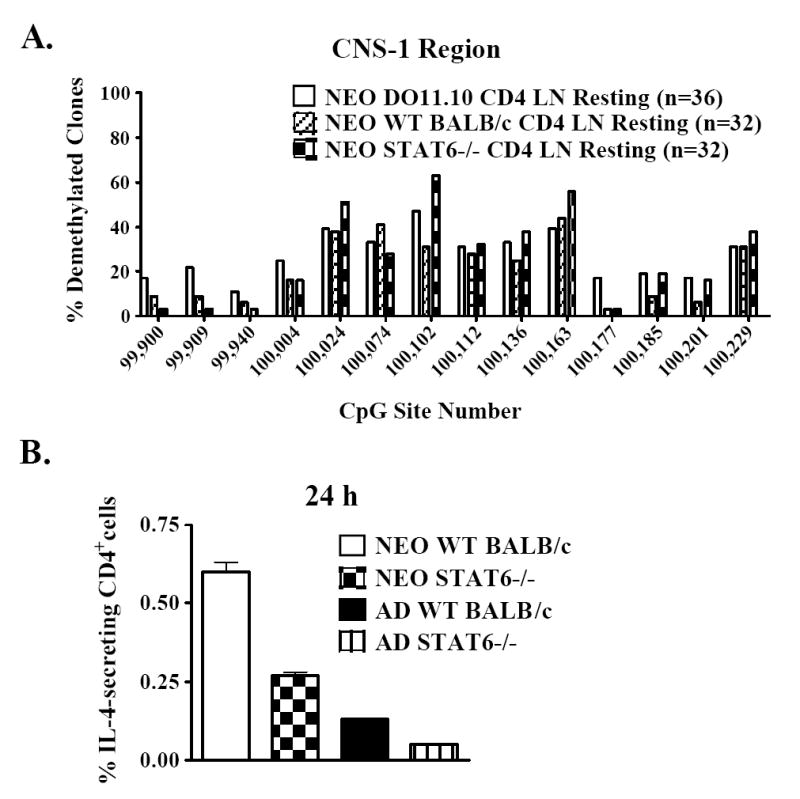
(a) CNS-1 methylation levels were assessed in resting CD4+ cells from day 7 neonatal DO11.10 TCR-transgenic, BALB/c wild-type (WT), or STAT6-deficient mice. (b) CD4+ cells from day 7 neonatal BALB/c wild-type (WT) or STAT6-deficient mice were polyclonally activated for 24 h. The frequencies of IL-4-secreting cells were enumerated by ELISPOT as described in Figure 2. The depicted experiment is representative of 4 independent experiments.
CNS-1 demethylation is already present in unactivated neonatal CD4 SP thymocytes, and this epigenetic state is greatly expanded in IL-4-producing cells
Around the time of birth, the first wave of mature CD4 SP thymocytes exits the thymus and colonizes the peripheral lymphoid compartments (34). Since resting neonatal peripheral CD4+ lymphocytes were already demethylated at CNS-1 (Figs. 6 and 8a), we next tested whether demethylation was established during T cell development in the thymus. CNS-1 methylation levels were assessed in resting CD4 SP thymocytes sorted from day 2 DO11.10 neonates. For most of the CNS-1 CpG sites, the levels of demethylation in day 2 neonatal CD4 SP thymocytes were similar to those of naïve resting CD4+ CD44lo lymph node cells from day 7 animals (Fig. 9a). These findings demonstrate that CNS-1 demethylation is initially patterned within the thymus during early life.
Figure 9. CNS-1 demethylation is already present in unactivated neonatal CD4 SP thymocytes, and this epigenetic state is greatly expanded in IL-4-producing cells.

(a) CNS-1 methylation levels were measured in resting naïve CD4+ CD44lo lymph node (LN) cells from day 7 DO11.10 neonates or from CD4 single positive thymocytes (SP THY) sorted from day 2 DO11.10 neonates. (b) CD4 SP thymocytes from day 3 neonatal DO11.10 mice were enriched and activated for 2 days with plate-bound anti-CDε plus soluble anti-CD28 mAb. After 2 h of restimulation with PMA and ionomycin, the cells were stained for IL-4 expression using Miltenyi IL-4 Cell Enrichment and Detection Kits, followed by staining with anti-CD4 mAb. CD4+IL-4+ cells were sorted and cell preparations from 3 independent experiments were pooled for CNS-1 methylation analysis. CpG methylation levels in resting CD4 SP thymocytes are presented for comparison.
We next investigated whether the capacity for IL-4 expression corresponds with a demethylated state at CNS-1. To test this hypothesis, CD4 SP thymocytes from day 3 neonatal DO11.10 mice were activated for 2 days. The cells were then stained for IL-4 expression using Miltenyi IL-4 Cell Enrichment and Detection Kits, and IL-4-producing CD4+ cells were sorted for bisulfite methylation analysis. IL-4-expressing cells were considerably more demethylated at CNS-1 compared to freshly isolated resting cells (Fig. 9b). This result strongly suggests that a derepressed epigenetic state at CNS-1 is permissive for IL-4 expression in neonatal cells.
DISCUSSION
Unlike naïve adult cells, murine neonatal CD4+ T cells demonstrate effector-like Th2 function early after polyclonal stimulation in vitro. While enhanced Th2 function in neonates is well documented, the molecular processes that contribute to this phenomenon have not previously been studied. In this report, we found that the early high-level IL-4 production arose from a small population of neonatal CD4+ cells, did not require cell cycle entry, and was relatively unaffected by treatment with a global demethylating agent. An analysis of CpG methylation patterns at the Th2 cytokine locus revealed that resting neonatal CD4+ cells, unlike resting adult cells, were demethylated at a highly conserved control region, termed CNS-1 (20). This differential pattern of methylation between resting naïve neonatal and adult CD4+ cells was unique to CNS-1 since methylation profiles were similar at other investigated regions of the Th2 cytokine locus. Enhanced Th2 function and greater CNS-1 demethylation was a stable property of neonatal cells, since the frequencies of IL-4-secretors and the levels of CNS-1 demethylation were both higher in 5-day neonatal Th2 effectors compared to adult Th2 cells. The transcription factor STAT6 was not required for establishing CNS-1 demethylation, and this epigenetic state was already present in neonatal CD4 SP thymocytes. Lastly, CNS-1 demethylation was strongly overrepresented in IL-4-expressing neonatal CD4 SP thymocytes, compared to unactivated cells. Taken together, these data indicate that neonatal CD4+ cells have several distinct properties that predispose them toward rapid, high-level Th2 function.
The poised state of neonatal CD4+ T cells contrasts sharply with that of naïve adult cells, which have been shown to undergo extensive proliferation and epigenetic remodeling of the Th2 cytokine locus over a period of several days to weeks before high-level Th2 cytokine expression is achieved (4). Neonatal CD4+ cells possess at least 3 distinctive properties that could predispose them toward greater early Th2 effector development and function. First, we have found that a key enhancer region of Th2 cytokine expression, CNS-1 (20), was completely demethylated in a subset of resting neonatal CD4+ cells, but not in adult cells. Since the degree of demethylation at CNS-1 correlates strongly with the frequencies of Th2 cells that make IL-4 (14), this epigenetic state could promote early Th2 cytokine production by neonatal cells. Second, unlike adult cells (5, 6), neonatal CD4+ cells produced high levels of IL-4 protein early after activation. Since IL-4 drives Th2 differentiation (35), this could rapidly initiate the development of neonatal Th2 cells. Third, our laboratory has previously shown that activated neonatal CD4+ cells have more rapid cell division kinetics compared to adult cells (23). Thus, neonatal CD4+ cells could rapidly produce high levels of IL-4, which may be followed by more rapid proliferation of the IL-4-secreting neonatal cells, compared to adult cells. Together, these unique properties may give neonatal cells a ‘head start’ over adult cells during early Th2 cell development.
Since CNS-1 was uniquely demethylated in resting neonatal CD4+ cells, it is tempting to speculate that this regulatory region alone is responsible for the differences in IL-4 and IL-13 production between recently activated neonatal and adult CD4+ cells. In transgenic adult mice, CNS-1 has been shown to be a coordinate regulator of human Il4 and Il13 gene expression (20) and a targeted deletion of the murine CNS-1 region disrupts Th2 differentiation in vitro and in vivo (36). It has been suggested that epigenetic modifications at this region are permissive for Il4 and Il13 gene transcription (37). We have demonstrated that two other regulatory regions of both Il4 and Il13 expression, rad50 RHS7 within the Th2 cytokine LCR (11), and CS-1 of the distal Il13 promoter (30), exhibited similar DNA methylation patterns in neonatal and adult cells. Thus, recently activated neonatal cells could initially employ a demethylated CNS-1 region to enhance their early Th2 cytokine expression. Other control regions of the locus may also become involved as neonatal cells differentiate into Th2 effectors, similar to what occurs in adult cells. This would explain why there are initially large differences in the frequencies of IL-4-secreting neonatal and adult CD4+ cells, but smaller differences at the Th2 effector stage.
How could differences in DNA methylation levels between resting neonatal and adult CD4+ cells give rise to differences in Il4 and Il13 expression between recently activated neonatal and adult cells? CNS-1 contains putative binding sites for a number of transcriptional activators, including CREB, NFAT, ATF, AP-1, and c-Myb, and these binding sites each contain CpG dinucleotides (38). CpG methylation interferes with the binding of each of these transcription factors to their target sequences (39-42). Therefore, methylated CpG sites within CNS-1 could disrupt the binding of transcriptional activators to this region. Since CNS-1 has been shown to strongly interact with both the Il4 and Il13 promoters (43), it is possible that a demethylated CNS-1 region enhances Il4 and Il13 expression by bringing transcriptional activators in close proximity to the Th2 cytokine promoters. CNS-1 is relatively demethylated in resting neonatal CD4+ cells, a state that could facilitate the binding of transcriptional activators, resulting in high levels of Il4 and Il13 expression early after activation. In contrast, naïve resting adult CD4+ cells exhibit high levels of CNS-1 methylation. Transcription factor binding could therefore be inhibited in adult cells, resulting in lower levels of Il4 and Il13 expression following activation.
It is worth noting that while CNS-1 demethylation may be indicative of an open, permissive gene structure, it is not sufficient, per se, for high-level Th2 cytokine expression by neonatal CD4+ cells. As has previously been described for adult Th2 clones (14), there was a discrepancy between the frequencies of neonatal cells that were completely demethylated at CNS-1 and the frequencies of cells that actually produced IL-4. This was true for both recently activated cells and for Th2 effectors. Thus, while CNS-1 demethylation may establish a competent state at the Il4/Il13 locus (37), it is not sufficient, by itself, for high-level Th2 cytokine production by neonatal CD4+ cells. Other factors, such as the local availability of transcriptional activators, are also likely to play a role.
In light of our present observations, it will be interesting to determine what factors are responsible for establishing the unique epigenetic profile of neonatal CD4+ cells. Early IL-4 production and CNS-1 demethylation in neonatal cells did not require STAT6. STAT6-independent IL-4 expression and modifications at the Th2 cytokine locus have often been attributed to its downstream mediator GATA-3 (44). GATA-3 could be important for patterning CNS-1 demethylation in wild-type neonatal cells, as it has been shown to bind within this region (38).
Reports on adult and neonatal thymocytes support the idea that GATA-3 could establish CNS-1 demethylation within the thymus during early life. The de novo DNA methyltransferase Dnmt3b has been shown to bind within the CNS-1 region in resting adult CD4 SP thymocytes (26). Following activation of adult CD4 SP cells, Dnmt3b is preferentially excluded from CNS-1, an event that is associated with a marked decline in CpG methylation levels at this region (26). It has been suggested that GATA-3 binding may obstruct the recruitment of DNA methyltransferases, leading to demethylation of CNS-1 (26). Since neonatal CD4 SP thymocytes are already substantially demethylated in the resting state, it is possible that GATA-3 blocks Dnmt3b binding during this stage of thymocyte maturation. It has previously been shown that neonatal CD4 SP thymocytes express higher levels of GATA-3 than do adult cells (45). If resting neonatal CD4 SP cells had higher levels of GATA-3 bound to CNS-1, this could hinder Dnmt3b binding and result in greater DNA demethylation compared to resting adult cells. Since GATA-3 is expressed at all stages of thymocyte maturation (46), a second possibility is that neonatal thymocytes become demethylated in response to activation signals received prior to the CD4 SP stage. Thus, exclusion of Dnmt3b from the CNS-1 region, and ensuing CNS-1 hypomethylation, could potentially be established at or before the CD4 SP stage of thymocyte development.
High-level IL-4 expression (47) and CNS-1 demethylation are characteristics of neonatal CD4 SP thymocytes that are retained in peripheral CD4+ lymphocytes. It is possible that processes occurring in the periphery, such as lymphopenia induced proliferation (LIP), act to sustain these properties of neonatal cells. At birth, neonates are in a state of relative lymphopenia compared to adult animals. During the first week of life, the peripheral lymphoid organs are seeded by mature CD4 SP thymic emigrants. A proportion of these cells undergo LIP (48), a process that is associated with the acquisition of effector function when adult CD4+ T cells are adoptively transferred into neonatal hosts (49). It is not known whether endogenously proliferating neonatal T cells are also capable of effector function. However, naïve phenotype neonatal T cells also showed substantial demethylation at CNS-1, suggesting that mechanisms other than LIP contribute to the epigenetically poised state. Nonetheless, LIP may help to solidify the Th2 state in neonates. Experiments to address these and other related hypotheses are ongoing in our laboratory.
It is very clear that epigenetic modifications are critical for regulating gene expression during development. However, well characterized examples, such as globin gene switching (50) and selective TCR Vγ usage (51), apply to genes that are non-mutually expressed in fetal and adult life. The developmental regulation of Il4 represents a unique strategy whereby epigenetic modifications modulate the frequency of expression of a gene that is expressed throughout life. The epigenetic regulation of cytokine expression in early life may be a common theme in mice and men. In human newborns, repressive gene structures are thought to be responsible for the reduced expression (compared to adults) of IFN-γ in cord blood CD4+ T cells (52) and IL-12 in cord blood dendritic cells (53). Thus, it appears that certain cytokine response patterns of neonatal immune cells may already be pre-programmed at birth, and that the epigenetic state of the cytokine genes promotes anti-inflammatory responses. Significant contributions from several laboratories now suggest that the Th2-biased immune responses of murine neonates arise through a combination of cell intrinsic ((3, 54) and the present manuscript) and environmental (55) factors that are unique to early life.
Acknowledgments
The authors are grateful to Dr. Marcela Nouzova and Dr. Matthew Pipkin for their insightful review of the data and for stimulating discussions and to Patricia Guevara, Vesna Jurecic, Roland Jurecic, and Jim Phillips for excellent technical assistance. The authors would also like to thank Dr. Christopher Wilson, Department of Immunology, University of Washington School of Medicine, for the sodium bisulfite conversion protocol and for insightful discussions regarding the manuscript.
This work was supported by NIH grant number RO1 AI44923-02 (b.a.) and NIH NRSA number 1F30 ES012850-01 (s.r.).
Nonstandard Abbreviations
- CNS-1
conserved non-coding 1
- CS-1
consensus sequence 1
- LCR
locus control region
- RHS
rad50 hypersensitivity site
- CpG
cytosine-guanine dinucleotide 5’ – 3’
- LIP
lymphopenia induced proliferation
- CD
cluster of differentiation
- SP
single positive
- Dnmt
DNA methyltransferase
- S
synthesis phase of the cell cycle
- G2
second growth phase of the cell cycle
- M
mitotic phase of the cell cycle
- Vγ
gamma gene segment of the antigen receptor
References
- 1.Adkins B, Leclerc C, Marshall-Clarke S. Neonatal adaptive immunity comes of age. Nat Rev Immunol. 2004;4:553–564. doi: 10.1038/nri1394. [DOI] [PubMed] [Google Scholar]
- 2.Adkins B, Hamilton K. Freshly isolated, murine neonatal T cells produce IL-4 in response to anti-CD3 stimulation. J Immunol. 1992;149:3448–3455. [PubMed] [Google Scholar]
- 3.Adkins B, Bu Y, Guevara P. Murine neonatal CD4+ lymph node cells are highly deficient in the development of antigen-specific Th1 function in adoptive adult hosts. J Immunol. 2002;169:4998–5004. doi: 10.4049/jimmunol.169.9.4998. [DOI] [PubMed] [Google Scholar]
- 4.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 5.Richter A, Lohning M, Radbruch A. Instruction for cytokine expression in T helper lymphocytes in relation to proliferation and cell cycle progression. J Exp Med. 1999;190:1439–1450. doi: 10.1084/jem.190.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 7.Ho IC, Glimcher LH. Transcription: tantalizing times for T cells. Cell. 2002;109(Suppl):S109–120. doi: 10.1016/s0092-8674(02)00705-5. [DOI] [PubMed] [Google Scholar]
- 8.Takemoto N, Koyano-Nakagawa N, Yokota T, Arai N, Miyatake S, Arai K. Th2-specific DNase I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int Immunol. 1998;10:1981–1985. doi: 10.1093/intimm/10.12.1981. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc Natl Acad Sci U S A. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 13.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 14.Guo L, Hu-Li J, Zhu J, Watson CJ, Difilippantonio MJ, Pannetier C, Paul WE. In TH2 cells the Il4 gene has a series of accessibility states associated with distinctive probabilities of IL-4 production. Proc Natl Acad Sci U S A. 2002;99:10623–10628. doi: 10.1073/pnas.162360199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 16.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 17.Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sozeri O, Lohning M, Hu-Li J, Niesner U, Kreher S, Friedrich B, Pannetier C, Grutz G, Walter J, Paul WE, Radbruch A. A critical control element for interleukin-4 memory expression in T helper lymphocytes. J Biol Chem. 2005 doi: 10.1074/jbc.M502038200. [DOI] [PubMed] [Google Scholar]
- 18.Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402–4406. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- 19.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 21.Adkins B, Bu Y, Cepero E, Perez R. Exclusive Th2 primary effector function in spleens but mixed Th1/Th2 function in lymph nodes of murine neonates. J Immunol. 2000;164:2347–2353. doi: 10.4049/jimmunol.164.5.2347. [DOI] [PubMed] [Google Scholar]
- 22.Adkins B, Du RQ. Newborn mice develop balanced Th1/Th2 primary effector responses in vivo but are biased to Th2 secondary responses. J Immunol. 1998;160:4217–4224. [PubMed] [Google Scholar]
- 23.Adkins B, Williamson T, Guevara P, Bu Y. Murine neonatal lymphocytes show rapid early cell cycle entry and cell division. J Immunol. 2003;170:4548–4556. doi: 10.4049/jimmunol.170.9.4548. [DOI] [PubMed] [Google Scholar]
- 24.Chakir H, Wang H, Lefebvre DE, Webb J, Scott FW. T-bet/GATA-3 ratio as a measure of the Th1/Th2 cytokine profile in mixed cell populations: predominant role of GATA-3. J Immunol Methods. 2003;278:157–169. doi: 10.1016/s0022-1759(03)00200-x. [DOI] [PubMed] [Google Scholar]
- 25.Baguet A, Bix M. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc Natl Acad Sci U S A. 2004;101:11410–11415. doi: 10.1073/pnas.0403334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol. 2003;4:1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 27.Cherwinski HM, Schumacher JH, Brown KD, Mosmann TR. Two types of mouse helper T cell clone. III. Further differences in lymphokine synthesis between Th1 and Th2 clones revealed by RNA hybridization, functionally monospecific bioassays, and monoclonal antibodies. J Exp Med. 1987;166:1229–1244. doi: 10.1084/jem.166.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaut M, Pierce JH, Watson CJ, Hanley-Hyde J, Nordan RP, Paul WE. Mast cell lines produce lymphokines in response to cross-linkage of Fc epsilon RI or to calcium ionophores. Nature. 1989;339:64–67. doi: 10.1038/339064a0. [DOI] [PubMed] [Google Scholar]
- 29.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 30.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol. 2001;167:4414–4420. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 31.Ho IC, Kaplan MH, Jackson-Grusby L, Glimcher LH, Grusby MJ. Marking IL-4-producing cells by knock-in of the IL-4 gene. Int Immunol. 1999;11:243–247. doi: 10.1093/intimm/11.2.243. [DOI] [PubMed] [Google Scholar]
- 32.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 34.Penit C, Vasseur F. Cell proliferation and differentiation in the fetal and early postnatal mouse thymus. J Immunol. 1989;142:3369–3377. [PubMed] [Google Scholar]
- 35.Perez VL, Lederer JA, Lichtman AH, Abbas AK. Stability of Th1 and Th2 populations. Int Immunol. 1995;7:869–875. doi: 10.1093/intimm/7.5.869. [DOI] [PubMed] [Google Scholar]
- 36.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, Locksley RM. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 37.Guo L, Hu-Li J, Paul WE. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity. 2005;23:89–99. doi: 10.1016/j.immuni.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Takemoto N, Kamogawa Y, Jun Lee H, Kurata H, Arai KI, O’Garra A, Arai N, Miyatake S. Cutting edge: chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for Th2-specific cytokine gene cluster. J Immunol. 2000;165:6687–6691. doi: 10.4049/jimmunol.165.12.6687. [DOI] [PubMed] [Google Scholar]
- 39.Cockerill PN, Bert AG, Jenkins F, Ryan GR, Shannon MF, Vadas MA. Human granulocyte-macrophage colony-stimulating factor enhancer function is associated with cooperative interactions between AP-1 and NFATp/c. Mol Cell Biol. 1995;15:2071–2079. doi: 10.1128/mcb.15.4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson CB, Wang CY, Ho IC, Bohjanen PR, Petryniak B, June CH, Miesfeldt S, Zhang L, Nabel GJ, Karpinski B, et al. cis-acting sequences required for inducible interleukin-2 enhancer function bind a novel Ets-related protein, Elf-1. Mol Cell Biol. 1992;12:1043–1053. doi: 10.1128/mcb.12.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klempnauer KH. Methylation-sensitive DNA binding by v-myb and c-myb proteins. Oncogene. 1993;8:111–115. [PubMed] [Google Scholar]
- 42.Penix LA, Sweetser MT, Weaver WM, Hoeffler JP, Kerppola TK, Wilson CB. The proximal regulatory element of the interferon-gamma promoter mediates selective expression in T cells. J Biol Chem. 1996;271:31964–31972. doi: 10.1074/jbc.271.50.31964. [DOI] [PubMed] [Google Scholar]
- 43.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 44.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 45.Koyanagi M, Imanishi K, Arimura Y, Kato H, Yagi J, Uchiyama T. Immunologic immaturity, but high IL-4 productivity, of murine neonatal thymic CD4 single-positive T cells in the last stage of maturation. Int Immunol. 2004;16:315–326. doi: 10.1093/intimm/dxh027. [DOI] [PubMed] [Google Scholar]
- 46.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 47.Bendelac A, Schwartz RH. CD4+ and CD8+ T cells acquire specific lymphokine secretion potentials during thymic maturation. Nature. 1991;353:68–71. doi: 10.1038/353068a0. [DOI] [PubMed] [Google Scholar]
- 48.Le Campion A, Bourgeois C, Lambolez F, Martin B, Leaument S, Dautigny N, Tanchot C, Penit C, Lucas B. Naive T cells proliferate strongly in neonatal mice in response to self-peptide/self-MHC complexes. Proc Natl Acad Sci U S A. 2002;99:4538–4543. doi: 10.1073/pnas.062621699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arai N, Nomura D, Villaret D, DeWaal Malefijt R, Seiki M, Yoshida M, Minoshima S, Fukuyama R, Maekawa M, Kudoh J, et al. Complete nucleotide sequence of the chromosomal gene for human IL-4 and its expression. J Immunol. 1989;142:274–282. [PubMed] [Google Scholar]
- 50.Harju S, McQueen KJ, Peterson KR. Chromatin structure and control of beta-like globin gene switching. Exp Biol Med (Maywood) 2002;227:683–700. doi: 10.1177/153537020222700902. [DOI] [PubMed] [Google Scholar]
- 51.Agata Y, Katakai T, Ye SK, Sugai M, Gonda H, Honjo T, Ikuta K, Shimizu A. Histone acetylation determines the developmentally regulated accessibility for T cell receptor gamma gene recombination. J Exp Med. 2001;193:873–880. doi: 10.1084/jem.193.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO- T cells. J Immunol. 2002;168:2820–2827. doi: 10.4049/jimmunol.168.6.2820. [DOI] [PubMed] [Google Scholar]
- 53.Goriely S, Van Lint C, Dadkhah R, Libin M, De Wit D, Demonte D, Willems F, Goldman M. A defect in nucleosome remodeling prevents IL-12(p35) gene transcription in neonatal dendritic cells. J Exp Med. 2004;199:1011–1016. doi: 10.1084/jem.20031272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Lee HH, Bell JJ, Gregg RK, Ellis JS, Gessner A, Zaghouani H. IL-4 utilizes an alternative receptor to drive apoptosis of Th1 cells and skews neonatal immunity toward Th2. Immunity. 2004;20:429–440. doi: 10.1016/s1074-7613(04)00072-x. [DOI] [PubMed] [Google Scholar]
- 55.Sun CM, Deriaud E, Leclerc C, Lo-Man R. Upon TLR9 Signaling, CD5(+) B Cells Control the IL-12-Dependent Th1-Priming Capacity of Neonatal DCs. Immunity. 2005;22:467–477. doi: 10.1016/j.immuni.2005.02.008. [DOI] [PubMed] [Google Scholar]



