Abstract
Little is known about the mechanisms that regulate species-specific telomere length, particularly in mammalian species. The genetic regulation of telomere length was therefore investigated by using two inter-fertile species of mice, which differ in their telomere length. Mus musculus (telomere length >25 kb) and Mus spretus (telomere length 5–15 kb) were used to generate F1 crosses and reciprocal backcrosses, which were then analyzed for regulation of telomere length. This analysis indicated that a dominant and trans-acting mechanism exists capable of extensive elongation of telomeres in somatic cells after fusion of parental germline cells with discrepant telomere lengths. A genome wide screen of interspecific crosses, using M. spretus as the recurrent parent, identified a 5-centimorgan region on distal chromosome 2 that predominantly controls the observed species-specific telomere length regulation. This locus is distinct from candidate genes encoding known telomere-binding proteins or telomerase components. These results demonstrate that an unidentified gene(s) mapped to distal chromosome 2 regulates telomere length in the mouse.
Telomeres are structures at the termini of eukaryotic chromosomes that appear to be important for maintaining the integrity of chromosomes, for chromosomal positioning in the nucleus, for transcriptional silencing, and for cellular replicative capacity (1–11). Although factors contributing to shortening or maintenance of telomere length have been identified (5, 8, 11–13), the mechanisms underlying overall regulation of telomere length in mammalian cells are not well defined. Telomere length is relatively well conserved within each mammalian species, but varies widely across species. Although genetic regulation of telomere length has been studied in yeast (14) and in maize (15), to date the genetic basis for telomere length polymorphism has not been assessed in mammalian species. In the present study, we therefore used a telomere length polymorphism between mice of inter-fertile species to analyze the genetic basis underlying telomere length differences.
MATERIALS AND METHODS
Mice.
BALB/c mice were obtained from Frederick Cancer Research Center, Frederick, MD. M. spretus were generously provided by Michael Potter (National Institutes of Health, Bethesda, MD) or purchased from The Jackson Laboratory. Interspecies F1 mice and backcross mice were generated and housed in the animal facility at PerImmune, Rockville, MD or Bioqual, Rockville, MD.
Telomere Length Analysis.
Single cell suspensions of spleen were generated by conventional methods. Single cell suspensions of liver were generated by incubating minced livers with collagenase, type IV, (2 μg/ml; Sigma) for 30 min at 37°C. After depleting red blood cells, cell suspensions were mixed with an equal volume of 1.2% Incert Agarose (FMC) to make plugs containing 5 × 105 cells. Plugs were treated with Proteinase K (1 μg/ml; Life Technologies, Grand Island, NY) for 18–20 hr at 45°C and subsequently incubated with phenylmethylsulfonyl fluoride (1 μg/ml; Sigma) for 1 hr at 37°C. DNA was digested by incubating the plugs with 100 units/ml each of HinfI and RsaI (Boehringer Mannheim) for 18–20 hr at 37o. Digested DNA was separated on a 1% Ultrapure agarose (Life Technlogies) gel by using a Bio-Rad CHEF MAPPER pulsed field gel electrophoresis apparatus at recommended conditions for separating 5–100 kb DNA. High molecular weight (48–8 kb) (Bio-Rad) and Lambda Hindi III (23–0.1 kb) (Stratagene) markers were included in all gels.
DNA was stained with ethidium bromide (Life Technologies) and photographed, and gels were dried at 65°C for 1.5 hr. Gels were denatured, neutralized, hybridized with a 32P end-labeled oligonucleotide (CCCTAA)4 probe (David Winkler, NIH), and analyzed by PhosphorImager (Molecular Dynamics).
Telomere length also was measured by quantitative fluorescence in situ hybridization as described (16). Spleen cells were stimulated in vitro for 48 hr with a mixture of 5 μg/ml Con A (Sigma), 15 μg/ml lipopolysaccharide (Sigma), and 100 units/ml human rIL2 (Chiron). Colcemid (10 μg/ml, GIBCO) was added for the final 30 min of culture. Cells were harvested, fixed with methanol/acetic acid, air dried on slides overnight, fixed with 4% formaldehyde, pepsin treated, dehydrated, and air dried for 2 hr at room temperature. Hybridization was then carried out with Cy-3-conjugated (C3TA2)3 peptide nucleic acid probe (PBIO/Biosearch, Bedford, MA) as reported (16). Digital images of metaphase chromosomes counterstained with 4′,6-diamidino-2-phenylindole were recorded with a MicroImager MI1400–12 camera (Xillix; Vancouver) on an Axioplan fluorescence microscope (Zeiss). Image analysis was performed with dedicated software to calculate integrated telomere fluorescence intensity, which was expressed in telomere fluorescence units with each unit corresponding to the fluorescence obtained from 1 kb of T2AG3 repeats in hybridized plasmid DNA (17). Details of these methods have been reported (16).
Chromosomal Mapping.
Linkage studies were performed by using high stringency Southern blot hybridizations or microsatellite PCRs as described (18). For the Terf1 gene, a PCR product (forward primer, bp 89–108; reverse primer, bp 410–391) from a mouse expressed sequence tag (EST) (GenBank accession no. AA103157) with 98% sequence identity over 351 bp to the mouse Terf1 was used as a probe on Southern blots. Similarly for the telomerase reverse transcriptase gene (Tert), a PCR product (forward T7 and reverse bp 248–265) from a human EST (GenBank accession no. AA281296) with 99% sequence identity over 389 bp with TERT (19, 20); for telomerase protein 1 (Tp1), a PCR product (T7T3) from a mouse EST (GenBank accession no. AA139221) with 100% sequence identity over 415 bp (with one gap) to mouse Tep1; and for Terf2, a PCR product (T7T3) from a human EST (GenBank accession no. AA163408) with 99% sequence similarity to Terf2 (21) over 302 bp were used as probes. (Note: for the latter EST, the greater sequence identity with mouse rather than human TERF2 indicates that this sequence is really a mouse rather then human EST). For each probe, informative restriction fragments that hybridized to single bands in each “strain” of mice segregated in the interspecific backcross were identified: For Terf1, BamHI fragments (C3H/HeJ-gld, 6.5 kb; M. spretus, 3.0 kb); for Tert, BglII fragments (C3H/HeJ-gld, 8.0 kb, 5.5 kb; M. spretus, 7.0 kb, 4.3 kb); for Tep1, BamHI fragments (C3H/HeJ-gld, 12 kb; M. spretus, 14 kb); and for Terf2, BglII fragments (C3H/HeJ-gld, 4.8 kb; M. spretus, 4.5 kb). Polymorphisms for Massachusetts Institute of Technology (MIT) microsatellite markers have been defined (http://www-genome.wi.mit.edu/cgi-bin/mouse/index).
RESULTS
Telomere Length in F1 Progeny of M. musculus and M. spretus.
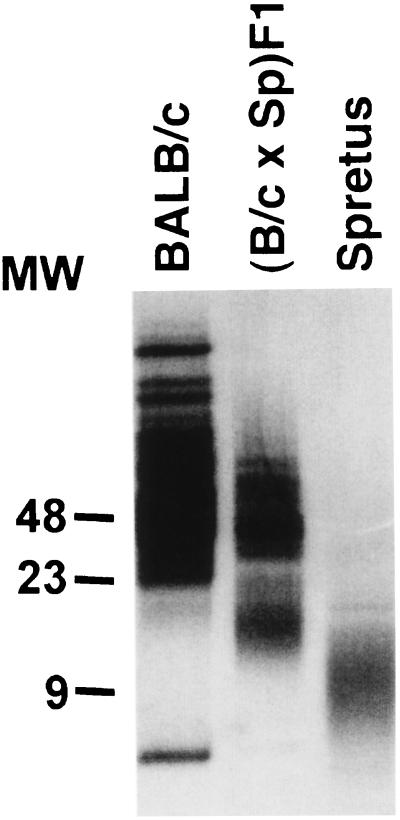
When telomere lengths in BALB/c (M. musculus) and M. spretus somatic cells were analyzed by pulsed field electrophoresis, BALB/c telomeres were significantly longer (>25 kb) than those from M. spretus (5–15 kb) (Fig. 1), consistent with previous reports (22, 23). The existence of multiple tracts in the long telomeres of BALB/c observed here is consistent with previous characterization of M. musculus telomeres (22). The contribution of species-specific factors to the determination of telomere length was analyzed in F1 crosses of inter-fertile BALB/c and M. spretus mice and revealed a bimodal distribution of telomere restriction fragment (TRF) length, consistent with the recent findings of Coviello-McLaughlin and Prowse (24). When this bimodal distribution was analyzed more closely, it was reproducibly observed that telomere restriction fragments from F1 mice consist of a longer set of telomeres corresponding to the length seen in BALB/c and a shorter set that is significantly longer than those in M. spretus (Fig. 1).
Figure 1.
Telomere length of BALB/c (M. musculus), spretus (M. spretus), and (BALB/c × M. spretus)F1 somatic cells. Genomic DNA was isolated from spleen cells of adult BALB/c, M. spretus, and F1 mice, subjected to restriction digestion, and analyzed by pulse field electrophoresis.
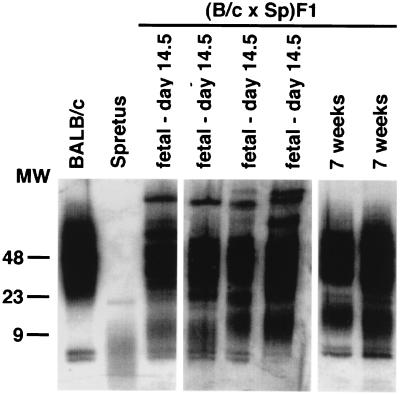
At the time of fertilization of BALB/c ova by M. spretus spermatozoa, chromosomes of each parental origin are present, presumably with telomeres of corresponding parental length. Because adult F1 mice were found to have TRF length distributions that did not include short TRF corresponding to the M. spretus phenotype, the possibility was tested that somatic cell telomeres of M. spretus origin increased in length during F1 development. TRF preparations from fetal and adult F1 donors all contained an equivalent set of long TRF, resembling the distribution seen in BALB/c (Fig. 2). In contrast, the set of lower molecular weight TRF was longer in day 14 fetal F1 preparations than in M. spretus parental DNA, with individual mice varying in the degree of lengthening. This lower set of telomere bands was lengthened to a more uniform degree by 7 wk of age. Telomere length in parental BALB/c and M. spretus somatic cells did not vary significantly over this age range (data not shown). The observed increases in the length of the shorter TRF band during ontogenic development of F1 mice suggests that a genetically dominant mechanism exists which is trans-active in its ability to mediate substantial lengthening of M. spretus origin telomeres during normal somatic cell development.
Figure 2.
Ontogeny of telomere length expression in (BALB/c × M. spretus)F1 somatic cells. Genomic DNA was isolated from liver, digested, and analyzed as described in Methods.
Although TRF measurements provide a widely used assay of telomere length, the restriction fragments detected by this method consist of a component of subtelomeric DNA in addition to telomeric repeats. Telomere length in BALB/c, M. spretus, and F1 spleen cells also was assessed therefore more directly by quantitative fluorescence hybridization (Q-FISH). When the distribution of fluorescence intensity of individual metaphase chromosomes was analyzed, a pattern was observed that very closely paralleled that seen by TRF analysis (Fig. 3). The estimated telomere length of M. spretus chromosomes was shorter than that of BALB/c chromosomes. Moreover, analysis of F1 telomeres revealed a broad bimodal fluorescence distribution, indicating that essentially all chromosomes have telomeres longer than those observed in M. spretus. These findings confirm the conclusion that telomeres on spretus origin chromosomes have been substantially lengthened during F1 somatic cell development.
Figure 3.
Quantitation of telomere length by fluorescence in situ hybridization. Spleen cells from M. spretus, BALB/c, or (BALB/c × M. spretus)F1 mice were stimulated in vitro. Metaphase preparations derived from these activated cells were analyzed by quantitative fluorescence hybridization with a Cy3-labeled telomere-specific peptide nucleic acid probe (16, 17). Telomere fluorescence intensity was calculated from digital images by using dedicated software and is expressed in telomere fluorescence units with each unit corresponding to 1 kb of hybridized T2AG3 in plasmid DNA (16) by in situ hybridization with a fluorescein isothiocyanate-conjugated telomere-specific peptide–nucleic acid probe, and telomere length was calculated by scanning and quantitation of fluorescence signal.
Determination of Telomere Length in Backcross Progeny.
To further analyze the role of genetic polymorphisms in determining telomere length, backcross analyses were carried out. The observation that telomeres in F1 mice tended to “lengthen” relative to the M. spretus parent suggested that the long BALB/c phenotype might be genetically dominant. Consistent with this hypothesis, TRF from all (BALB/c × M. spretus)F1 × BALB/c backcross mice consisted of a population of longer fragments equivalent to the length of BALB/c TRF and a population of shorter TRF, which were longer than those in M. spretus and were in fact longer than the population of shorter TRF observed in F1 mice (Fig. 4A).
Figure 4.
Telomere length in F1 × spretus backcross mice. Genomic DNA was isolated from spleen cells of adult backcross mice, subjected to restriction digestion, and analyzed by pulse field electrophoresis. (A) Telomere length in (BALB/c × M. spretus)F1 × BALB/c backcross mice. The numbers indicated for F1 and backcross mice identify individual animals. (B) Telomere length in (BALB/c × M. spretus)F1 × M. spretus backcross mice. Numbers identify individual backcross mice.
If the M. spretus phenotype were recessive, offspring of the F1 × M. spretus backcross might reveal a segregation of TRF phenotypes, reflecting genetic segregation of the gene(s) that determine telomere length. When this backcross was analyzed, a segregation of telomere length phenotypes was indeed observed. All offspring had a bimodal TRF distribution, with a uniform larger band that was once again equivalent to the BALB/c distribution (Fig. 4B). There was, however, substantial length polymorphism in the shorter TRF bands of these backcross mice. In an initial analysis, five of 31 offspring analyzed expressed a TRF band that was shorter than that seen in F1 mice and was equivalent to that in M. spretus parental mice. The remaining backcross mice expressed TRF similar to those in F1 × M. musculus backcross mice or showed an apparently intermediate phenotype. This segregation of the short or nonlengthening TRF phenotype was consistent with the influence of a gene or genes that are polymorphic in M. musculus and M. spretus and for which the spretus or shorter telomere phenotype is recessive.
Mapping of Polymorphic Genetic Loci Involved in Determination of Telomere Length.
In an extended segregation analysis, candidate genes corresponding to telomere- or telomerase-associated genes were first considered. The chromosomal positions of four candidate genes in the mouse were defined using another interspecific backcross DNA panel that allows precise mapping of genes throughout the mouse genome (Fig. 5A). The gene encoding mouse telomere repeat-binding factor (Terf1)(25) was mapped to proximal mouse chromosome 1 at consensus map position 11.8 cM. In addition, the recently cloned telomere repeat-binding factor 2 gene (Terf2) (21) was mapped to mouse chromosome 8 at consensus map position 50 cM, the telomomerase reverse transcriptase gene (Tert) (19, 20) to mouse chromosome 13 at consensus map position 48 cM, and the telomere protein (Tep1)(26) gene to mouse chromosome 14 at consensus map position 18.9 cM. Microsatellites corresponding to these chromosmal positions were then examined in mice characterized for telomere length (Fig. 5B). None of these candidates were linked to telomere phenotype as assessed by a χ2 analysis (P > 0.5).
Figure 5.
Examination of candidate genes for telomere length regulation. (A) Chromosomal localization of Terf1, Terf2, Tert, and Tep1. The segregation of these genes among flanking genetic loci on mouse chromosomes 1, 8, 13, and 14 in [(C3H/HeJ-gld × M. spretus) F1 × C3H/HeJ-gld] interspecific backcross mice is shown. Filled boxes represent the homozygous C3H pattern and open boxes represent the F1 pattern. The number of mice with each haplotype is shown in each column. The mapping of the reference loci as well as the relationship to other mouse chromosomal markers is available on the internet: http://www.informatics.jax.org/crossdata.html to enter DNA Mapping Panel Data Sets from MGI (Mouse Genome Informatics), then select the Seldin cross and Chromosome. The consensus map positions derived from interpolation on the most recent composite maps (http://www.informatics.jax.org/bin/ccr/current/index) indicate the following map positions: Terf1 at 11.8 cM on mouse chromosome 1, Terf2 at 50 cM on mouse chromosome 8, Tert at 49 cM on mouse chromosome 13, and Tep1 at 18.9 cM on mouse chromosome 14. (B) χ2 analysis of genotyping results of microsatellite markers for candidate genes and chromosomal regions. The genotypes of long and short telomere mice are shown (SS, homozygous M. spretus genotype; BS, heterozygous BALB/c and M. spretus genotype or C57BL/6J and M. spretus genotype). Note that the results for the X-chromosome show segregation distortion or pseudo-segregation distortion consistent with other studies in similar interspecies crosses (28). (C) Haplotype analysis of distal mouse chromosome 2 in 28 short telomere mice. Each column corresponds to the observed haplotype pattern with the typing results for each marker indicated by either filled (homozygous M. spretus) or open (heterozygous genotype) boxes. The number of mice observed with each haplotype is shown at the bottom of each column. The approximate chromosomal position of each microsatellite marker on each chromosome based on the most recent consensus map is shown adjacent to each marker.
A genome wide search for candidate loci that might be involved in telomere length determination was initiated by analysis of microsatellite chromosomal markers in an extended series of 93 backcross mice. If M. spretus homozygosity at a locus is necessary for expression of the short telomere phenotype, then candidate loci are those which are of homozygous M. spretus (SS) origin in each of the F1 × M. spretus backcross mice with that phenotype. Ten backcross mice and parental controls were analyzed for polymorphic markers at ≈15-cM intervals. Because positive interference is strong in mouse meiotic recombination, double crossovers within a 15 cM interval are nearly excluded (18), and there is high confidence in exclusion of chromosomal regions. These studies involving the analysis of >120 microsatellite markers identified two large chromosomal regions (on mouse chromosomes 2 and X) that were not excluded using this simple paradigm. Subsequent studies focusing on these two regions provided very strong support for a locus on distal mouse chromosome 2 (Fig. 5B). Each of 28 short telomere backcross mice were homozygous for the M. spretus genotype at D2Mit74, and all of the 53 mice that had the heterozygous genotype at this locus (SB genotype) had the long telomere phenotype. Considering only the 28 short telomere phenotype mice, the chances of observing only the homozygous M. spretus genotype at this locus in a genome wide analysis is <10−10. The P values for the χ2 analysis (Fig. 5B) and the analysis of the short telomere mice alone are both several orders of magnitude beyond those values considered to be highly significant for linkage of a genetic marker with a phenotype for a complex genetic trait (27).
The observation that 12 of 40 mice that were homozygous for the M. spretus genotype at this locus had the long telomere phenotype indicates that this locus cannot account for all of the genetic control of telomere length in these crosses. Additional microsatellites on distal mouse chromosome 2 were examined in the short telomere phenotype mice to define more precisely the critical genetic interval. These studies indicate that the putative gene for this phenomenon most likely is located in the distal 5 cM of mouse chromosome 2 (Fig. 5C).
DISCUSSION
The mechanisms involved in regulation of telomere length are not well characterized. Although studies in yeast (14) and in maize (15) have suggested that telomere length in these species is under polygenic control, no more precise analysis of these genetic influences has been reported, and there has been no previous evaluation of genetic regulation of telomere length in mammalian species.
When M. musculus BALB/c (long telomeres) and M. spretus (short telomeres) mice were interbred, the resulting F1 offspring expressed a pattern of telomere length that was different from that expected if both parental sets of chromosomes had maintained the telomere length characteristic of their species of origin. F1 chromosomes uniformly expressed telomeres with a bimodal size distribution on pulsed field gel analysis or by in situ hybridization. One set of telomeres in F1 mice was similar to that of the M. musculus parent. However, a second set of telomeres, although significantly shorter than this first set, was substantially longer than those observed in M. spretus parental mice. The fertilized egg resulting from M. musculus × M. spretus breeding presumably contains two sets of parental chromosomes, each with telomeres of a length corresponding to that of the parental species. The shorter set of telomeres observed in F1 mice is likely to be of M. spretus origin, although this interpretation has not been directly demonstrated. The fact that these shortest F1 telomeres are significantly longer than those in M. spretus indicated that a substantial increase in telomere length occurred during normal embryonic, fetal, and/or postnatal somatic cell development. This increase represents a unique and potentially significant demonstration that substantial lengthening of telomeres can occur during normal somatic cell development. Although the functional consequences of this telomere lengthening are not clear, the existence of a mechanism for extending telomere length in somatic cells may provide a mechanism for extending replicative potential in circumstances in which this is important for physiologic function. It is of interest that a recent analysis of telomere length in populations of normal human B lymphocytes suggests that telomere length may increase during differentiation of mature naive B cells into germinal center B cells, which are in turn the precursors of long-lived memory B cells (29). In such a circumstance, telomere lengthening may serve to preserve and extend the capacity for clonal expansion that is critical to immune function and memory.
Backcrossing of F1 mice to the M. musculus BALB/c parent generated results consistent with a dominant determination of the long telomere phenotype, with further lengthening of the shorter TRF band beyond that seen in the F1. Most informative, however, was the outcome of backcrossing F1 mice to the M. spretus parent, in which offspring exhibited heterogeneous telomere length phenotypes consistent with the segregation of two or three unlinked loci, with M. spretus phenotype at all loci being required for the short or nonlengthening phenotype. The difficulty in assessing limited changes in length of the longer musculus telomeres precludes excluding co-dominant inheritance by which M. musculus origin telomeres might become shorter in an F1 cellular environment.
Genotype mapping of the short telomere (M. musculus × M. spretus) × M. spretus backcross mice excluded the chromosomal regions containing the recently mapped Tep1 gene (30) (encoding a telomerase protein component) (26) and previously unmapped Tert (encoding the telomerase reverse transcriptase) (19, 20), Terf1 (25) and Terf2 (21). Notably, the position of Terf1 in mouse is disparate from that previously reported (31) and is consistent with that predicted from the human chromosomal position reported on 8q12 (ref. 32 and for homology relationships see http://www.ncbi.nlm.nih.gov/Homology). In contrast, definitive mapping of a locus on distal chromosome 2 was evident from analysis of both the short and long telomere mice in this backcross. A small segment of 5 cM appears to contain an unknown gene that is responsible for most of the telomere length regulation reported here. All of the 28 short telomere mice were homozygous for the M. spretus genotype in this interval. Examining all of the long telomere mice indicated that most (53 of 65 mice) contained the heterozygous (M. musculus × M. spretus) genotype for this small chromosomal region. Because 28 of 40 mice with the homozygous M. spretus genotype had the short telomere phenotype, it is likely that this chromosomal region contains the predominant genetic determinant of telomere length polymorphism. At present, it is unclear how many additional genes might contribute to the fact that 12 of the 40 mice that are homozygous for the M. spretus genotype on distal chromosome 2 have the long telomere phenotype. No apparent candidate genes or ESTs have been identified in the mapped interval of mouse chromosome 2 or in the homologous region of the long arm of human chromosome 20 (20q13.2–13.3). Additional crosses are in progress to further narrow the interval for positional cloning of the putative gene responsible for determination of species-specific telomere length. This approach may have substantial implications with respect to understanding telomere length regulation in mammalian cells and conditions including malignancy and aging, in which such function may be important in determining tumor cell survival or longevity.
The mechanism(s) that mediates telomere lengthening in somatic cells are unclear. The most extensively studied mechanism of telomere extension in mammalian cells is that mediated by telomerase (12). It will be of interest to determine whether the telomere lengthening observed during somatic differentiation of M. musculus × M. spretus F1 mice is dependent on telomerase activity. This question should be subject to direct assessment with the recent development of telomerase-deficient mice by homologous recombination (33, 34). Alternative telomerase-independent mechanisms of telomere extension, which might involve processes such as unequal recombination, have been studied in single cell organisms (13) and also have been discussed as possible mechanisms for mammalian telomere regulation. The experimental approach undertaken in the present study should lead to the identification of a gene involved in telomere length regulation and may lead to further elucidation of the molecular mechanisms involved in control of mammalian telomere length.
Acknowledgments
We would like to thank Genevieve Sanchez at Bioqual, Rockville, MD for excellent animal care. We also would like to thank Drs. Michael Kuehn and Nan-Ping Weng for helpful discussions and critical review of this manuscript.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: cM, centimorgan; TRF, telomere restriction fragment; EST, expressed sequence tag.
References
- 1.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Greider C W. Curr Opin Genet Dev. 1994;4:203–211. doi: 10.1016/s0959-437x(05)80046-2. [DOI] [PubMed] [Google Scholar]
- 3.Harley C B, Villeponteau B. Curr Opin Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 4.Zakian V A. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 5.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 6.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 7.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaziri H, Schachter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley C B. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- 9.Vaziri H, Dragowska W, Allsopp R C, Thomas T E, Harley C B, Lansdorp P M. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang E, Harley C B. Proc Natl Acad Sci USA. 1995;92:11190–11194. doi: 10.1073/pnas.92.24.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng N, Levine B L, June C H, Hodes R J. Proc Natl Acad Sci USA. 1995;92:11091–11094. doi: 10.1073/pnas.92.24.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackburn E H. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 13.Lundblad V, Blackburn E H. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 14.Walmsley R M, Petes T D. Proc Natl Acad Sci USA. 1985;82:506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burr B, Burr F A, Matz E C, Romero-Severson J. Plant Cell. 1992;4:953–960. doi: 10.1105/tpc.4.8.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zijlmans J M, Martens U M, Poon S S, Raap A K, Tanke H J, Ward R K, Lansdorp P M. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martens U M, Zijlmans J M, Poon S S, Dragowska W, Yui J, Chavez E A, Ward R K, Lansdorp P M. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- 18.Watson M L, Seldin M F. Methods Mol Genet. 1994;5:369–384. [Google Scholar]
- 19.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 21.Broccoli D, Smogorzewska A, Chong L, de Lange T. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 22.Kipling D, Cooke H J. Nature (London) 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 23.Starling J A, Maule J, Hastie N D, Allshire R C. Nucleic Acids Res. 1991;18:6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coviello-McLaughlin G M, Prowse K R. Nucleic Acids Res. 1997;25:3051–3058. doi: 10.1093/nar/25.15.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 26.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 27.Lander E S, Schork N J. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 28.Montagutelli X, Tyrner R, Nadeau J H. Genetics. 1996;143:1729–1752. doi: 10.1093/genetics/143.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng N, Granger L, Hodes R J. Proc Natl Acad Sci USA. 1997;94:10827–10832. doi: 10.1073/pnas.94.20.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito T, Matsuda Y, Suzuki T, Hayashi A, Yuan X, Saito M, Nakayama J, Hori T. Genomics. 1997;46:46–50. doi: 10.1006/geno.1997.5005. [DOI] [PubMed] [Google Scholar]
- 31.Broccoli D, Chong L, Oelmann S, Fernald A A, Marziliano N, Vansteensel B, Kipling D, Lebeau M M, de Lange T. Hum Mol Genet. 1997;6:69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- 32.DeBry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 33.Blasco M A, Funk W, Villeponteau B, Greider C W. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- 34.Blasco M A, Lee H, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]