Abstract
Genetically engineered mouse models are powerful tools for studying cancer genes and validating targets for cancer therapy. We previously used a mouse lymphoma model to demonstrate that the translation initiation factor eIF4E is a potent oncogene in vivo. Using the same model, we now show that the oncogenic activity of eIF4E correlates with its ability to activate translation and become phosphorylated on Ser 209. Furthermore, constitutively activated MNK1, an eIF4E Ser 209 kinase, promotes tumorigenesis in a manner similar to eIF4E, and a dominant-negative MNK mutant inhibits the in vivo proliferation of tumor cells driven by mutations that deregulate translation. Phosphorylated eIF4E promotes tumorigenesis primarily by suppressing apoptosis and, accordingly, the anti-apoptotic protein Mcl-1 is one target of both phospho-eIF4E and MNK1 that contributes to tumor formation. Our results provide insight into how eIF4E contributes to tumorigenesis and pinpoint a level of translational control that may be suitable for therapeutic intervention.
[Keywords: Mouse model, tumorigenesis, apoptosis, translation, signaling]
Mutations that deregulate mRNA translation are common events in human cancers. For example, activation of phosphoinositol-3 kinase (PI3K) or a AKT/PKB, or inactivation of Pten, Tsc1, or Tsc2, increase translation of a subset of mRNAs that influence cell survival (for review, see Bilanges and Stokoe 2007). While each of these lesions affects additional processes, enforced expression of certain translation factors transforms rodent fibroblasts and promotes tumorigenesis (Lazaris-Karatzas et al. 1990; Ruggero et al. 2004; Wendel et al. 2004). Similarly, small molecules that target these pathways suppress protein translation and have anti-tumor effects (Dilling et al. 1994; Wendel et al. 2004). Thus, deregulation of translation is an important step in oncogenic transformation and may contribute to tumor maintenance.
Most protein translation involves assembly of the eIF4F translation initiation complex on the 5′ cap structure, followed by recruitment of ribosomal subunits and their associated factors (for review, see Hay and Sonenberg 2004). The eukaryotic translation initiation factor eIF4E contributes to the eIF4F complex, and its activity is controlled by mammalian target of rapamycin (mTOR) through phosphorylation of the eIF4E-binding proteins (4E-BP). mTOR-mediated phosphorylation of 4E-BP liberates eIF4E allowing it to enter the eIF4F complex. Although eIF4E is a general translation factor, it preferentially enhances translation of a subset of mRNAs with complex 5′ untranslated regions (UTRs). These include a number of transformation-related and survival genes (Rajasekhar et al. 2003; Mamane et al. 2007).
eIF4E is overexpressed in several tumors and has been linked to patient prognosis (for review, see Ruggero and Pandolfi 2003). Although the molecular basis underlying these associations is unknown, eIF4E affects cancer-relevant processes such as apoptosis (Polunovsky et al. 2000) and senescence (Ruggero et al. 2004), and acts as an oncogene alone or in combination with c-Myc (Lazaris-Karatzas et al. 1990; Ruggero et al. 2004; Wendel et al. 2004). Presumably, these functions reflect the ability of eIF4E to affect translation although other eIF4E-related functions may be required (Ruggero and Pandolfi 2003).
eIF4E is a target of PI3K and mitogen-activated protein kinase (MAPK) signaling, and may act as a convergence point for these pathways. Thus, the PI3K pathway leads to enhanced eIF4E activity via release from the 4E-BPs and ERK or p38/MAPK can increase phosphorylation of eIF4E via MNK1/2 (Waskiewicz et al. 1997). However, the biological significance of eIF4E phosphorylation and its effect on translation is controversial (for review, see Scheper and Proud 2002). In Drosophila eIF4E phosphorylation seems important for normal development (Lachance et al. 2002); mice with deletions in both Mnk kinases develop normally without detectable eIF4E phosphorylation (Ueda et al. 2004).
We previously used adoptive transfer methods to show that eIF4E is a potent oncogene in the Eμ-Myc transgenic mouse lymphoma model, producing aggressive and drug resistant tumors (Wendel et al. 2004). Here, we use the same model to further study eIF4E promotes tumorigenesis, identifying a regulatory role for the MNK kinases and one downstream effector, Mcl-1. These studies underscore the importance of eIF4E phosphorylation during oncogenesis, and identify a point of translational regulation that is a potential therapeutic target.
Results and Discussion
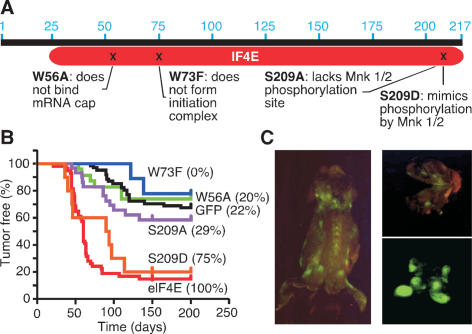
We used previously characterized eIF4E mutants to study its action during oncogenesis (Fig. 1A). The W56A mutant cannot bind the 5′ mRNA cap structure blocking translation initiation and nuclear–cytoplasmic export; the W73F mutation affects incorporation into the eIF4F translation initiation complex but not mRNA transport (Marcotrigiano et al. 1997); the S209 mutations target a conserved phosphorylation site for MNK1/2 kinases, with the serine-to-alanine (S209A) mutation abolishing phosphorylation and the serine-to-aspartic acid (S209D) mutation resembling phosphorylated eIF4E (Waskiewicz et al. 1997). These mutants were cloned into MSCV-IRES-GFP, and their expression verified in 3T3 cells following retroviral transduction (Supplementary Fig. 1A–C). All mutants were efficiently expressed, except W73A, which may be more prone to proteasomal degradation (Murata and Shimotohno 2006) (Supplementary Fig. 1C).
Figure 1.
eIF4E correlates with oncogenicity in vivo. (A) The eIF4 mutants used in this study. (B) Kaplan-Meier plot showing tumor onset data for mice reconstituted with Eμ-Myc HSCs expressing eIF4E (red, n = 45), or mutant alleles (W56A: green, n = 23; W73F: blue, n = 9; S209A: violet, n = 29; S209D: orange, n = 10) and vector (black, n = 61, includes historic controls). Numbers in brackets indicate the percentage of GFP-expressing tumors. (C) Fluorescence imaging of an Eμ-Myc/eIF4E tumor-bearing mouse (left), cerebral tumor (top right), and isolated tumors (bottom right).
We examined the ability of each eIF4E variant to cooperate with c-Myc during lymphomagenesis using a rapid adoptive transfer strategy as previously described for wild-type eIF4E (Wendel et al. 2004). Retroviruses encoding eIF4E or the various mutants were introduced into hematopoietic stem cells (HSCs) and used to reconstitute irradiated syngeneic recipients (Supplementary Fig. 2). These mice were monitored for lymphoma, and the tumor cells were analyzed for GFP expression.
As expected, wild-type eIF4E markedly accelerated tumor development (eIF4E vs. GFP: P < 0.0001) (Fig. 1B). By contrast, tumors arising from Eμ-Myc transgenic HSCs infected with the W56A or W73F alleles arose with a latency that resembled controls (W56A vs. GFP: P = 0.56; W73F vs. GFP: P = 0.46). Although only ∼25% the initial stem/progenitor cells were infected (data not shown), lymphomas arising from eIF4E-expressing HSCs were invariably GFP positive (Fig. 1B,C). HSCs infected with the other vectors were infected at a similar efficiency, but only 0%–22% of the resulting tumors were GFP positive, indicating they were not advantageous in vivo (Fig. 1B,C). Since the W56A mutant is defective in cap-dependent translation, these data imply that this function is crucial for eIF4E-mediated oncogenesis. The data also suggest that the mRNA transport functions are dispensable, although it remains possible that the reduced expression of the W73F mutant (also seen for the W73A mutant) contributes to the defect (Supplementary Fig. 2C).
The most surprising results arose from mice reconstituted with cells expressing eIF4E mutants affecting the MNK1/2 phosphorylation site. Mice reconstituted with cells expressing the S209A mutant were defective at promoting tumorigenesis (S209A vs. GFP: P = 0.24); by contrast, mice reconstituted with cells expressing the phosphomimetic S209D mutant displayed accelerated tumor onset comparable with wild-type eIF4E (S209D vs. GFP: P < 0.0001) (Fig. 1B). Seventy-five percent of the tumors arising from HSCs transduced with the S209D mutant were GFP positive, while only 28% of HSCs infected with S209A gave rise to tumors expressing the GFP reporter (Fig. 1B). Apparently, eIF4E phosphorylation at S209 of is important for oncogenicity.
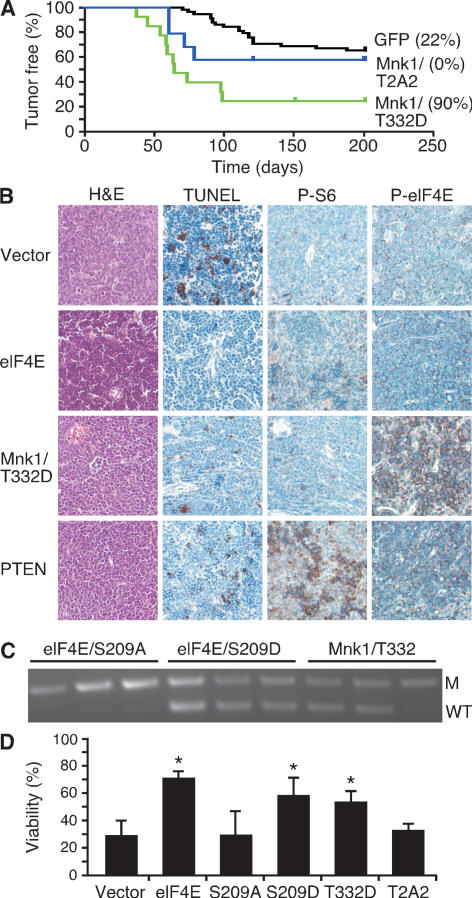
If eIF4E phosphorylation at Ser 209 is essential for oncogenesis, then constitutively activated MNK kinases might also be tumorigenic. We therefore examined the ability of an activated (T332D) or kinase-dead (T2A2) form of MNK1 (Waskiewicz et al. 1999) to promote tumorigenesis in the Eμ-Myc system. Like eIF4E, activated MNK1, but not the kinase-dead mutant, substantially accelerated lymphomagenesis (Mnk1 vs. GFP: P < 0.0001) (Fig. 2A), producing lymphomas with a mature B cell phenotype (Supplementary Table 1; Wendel et al. 2004). Both Mnk1- and eIF4E-expressing lymphomas showed low levels of apoptosis relative to most control tumors (Fig. 2B). Thus, MNK1 promotes tumor formation in a manner comparable with its substrate eIF4E.
Figure 2.
Activated MNK1 promotes lymphomagenesis. (A) Kaplan-Meier analysis of mice reconstituted with Eμ-Myc HSCs expressing an activated mutant Mnk1 (T332D) (green, n = 14), dominant-negative Mnk1 (T2A2) (blue, n = 10), or control (black, n = 61, controls as in Fig. 1). Numbers in brackets indicate the percentage of GFP-expressing tumors. (B) Representative micrographs of the indicated lymphomas stained as indicated. (C) Allele-specific PCR to detect the wild-type p53 (WT) and mutant allele (M) in tumors derived from Eμ-Myc/p53+/− HSCs transduced with the S209A and S209D mutants of eIF4E or an activated Mnk1 (T332D), and FACS sorted for GFP-expressing cells. (D) Viability of wild-type MEFs expressing c-Myc and empty vector or the indicated alleles and withdrawn from serum for 24 h. (*) Denotes P ∼ 0.02 compared with vector.
Disruption of apoptosis is a key event during Myc-induced lymphomagenesis (Strasser et al. 1990; Schmitt et al. 2002), and genes that suppress apoptosis, like eIF4E, Akt, or Bcl-2 can prevent p53 loss in lymphomas derived from p53+/− cells (Schmitt et al. 2002; Wendel et al. 2004). The observation that activated MNK1 also produces rapid tumors with a low apoptotic rate suggests that eIF4E phosphorylation attenuates apoptosis. To test this, we introduced mutant forms of eIF4E or Mnk1 into Eμ-Myc/p53+/− HSCs and determined whether the resulting tumors retained the wild-type p53 allele. Here, only tumors expressing the S209D mutant or activated MNK1—but not those expressing S209A—retained the wild-type p53 allele (three out of three for eIF4E S209D, and two out of three Mnk1) (Fig. 2C).
In parallel, we tested the ability of activated (T332D) and dominant-negative (T2A2) MNK1 and different eIF4E mutants to oppose c-Myc-induced apoptosis in mouse embryo fibroblasts (MEFs) (Evan et al. 1992). MEFs coinfected with c-Myc and eIF4E, its S209D mutant, or activated Mnk1 were resistant to apoptosis; in contrast, cells expressing vector alone, Mnk (T2A2), or the S209A mutant alleles of eIF4E remained sensitive (Fig. 2D). Thus, the oncogenic function of Mnk1 and eIF4E correlates with their ability to attenuate apoptosis in vitro and in vivo.
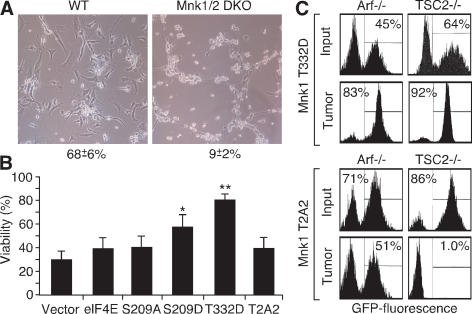
Our results linking Mnk to oncogenesis are surprising given that mice deficient in Mnk kinase develop normally (Ueda et al. 2004). Next, we tested whether MNK inhibition would sensitize cells to apoptosis in vitro or inhibit tumor expansion in vivo. MEFs derived from Mnk1/2 double-knockout (DKO) mice were transduced with Myc and examined for viability after serum depletion (Evan et al. 1992). Mnk1/2 DKO MEFs expressing Myc were hypersensitized to apoptosis (Fig. 3A, P < 0.0001) in a manner was reversed by reintroduction of the activated Mnk1 allele (T332D). However, in contrast to wild-type cells, Mnk1/2 DKO cells overexpressing both eIF4E and the S209A mutant remained hypersensitive to apoptosis (Fig. 3B).
Figure 3.
Activated MNK1 opposes Myc-induced apoptosis. (A) Phase contrast of c-myc-expressing wild-type (WT) and Mnk1/2 DKO (Mnk1/2 DKO) MEFs 16 h after serum withdrawal. Mean and standard deviation (n = 3) of cell viability are indicated. (B) Viability of Mnk1/2 DKO MEFs expressing c-Myc and vector or the indicated genes following serum withdrawal for 12 h. (*) P < 0.05; (**) P < 0.01. (C) Flow cytometry of tumor cell populations from Eμ-Myc/Arf−/− or Eμ-Myc/TSC2−/− lymphomas, partially transduced retroviruses coexpressing GFP, and constitutively active Mnk1 (T332D, top panels) or dominant-negative Mnk1 (T2A2, bottom panels) before (Input) and after (Tumor) lymphoma expansion in vivo.
We also examined tumor expansion of lymphoma cells expressing a dominant-negative MNK1 kinase mutant capable of attenuating eIF4e phosphorylation (T197A/T202A, denoted T2A2; see Supplementary Fig. 3; Waskiewicz et al. 1999). A subset of tumor cells was transduced with a Mnk-expressing MSCV-IRES-GFP-based vector and the mixed population transplanted into recipient animals. In this assay, an increase or decrease in GFP indicates that cells expressing the transgene have a selective advantage or disadvantage compared with controls, respectively. The assay was performed in Eμ-Myc/Arf−/− and Eμ-Myc/Tsc2−/− tumor cells, since TSC2-null lymphomas activate the mTOR pathway and ARF-null lymphomas do not (A. Malina and J. Pelletier, unpubl.) (Wendel et al. 2006).
The impact of Mnk inhibition was genotype dependent. The activated Mnk1 (T332D) conferred an advantage in vivo, and cells of either genotype expressing this mutant were enriched during tumor formation (Fig. 3C, top panels). By contrast, while the Mnk1 (T2A2) mutant had minimal effect in Arf−/− lymphoma cells, it dramatically inhibited the proliferation of Tsc2−/− cells, such that T2A2-expressing lymphoma cells were almost completely depleted from the population during lymphoma expansion (Fig. 3C, bottom panels). Thus, Mnk kinase inhibition appears to selectively inhibit tumor cells with deregulated translational control.
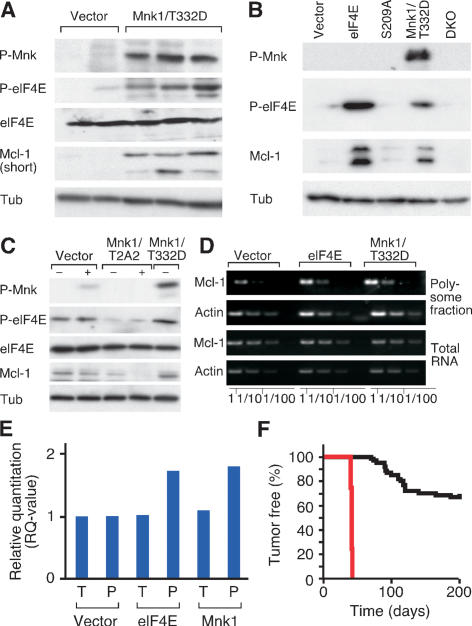
One potential anti-apoptotic target of phosphorylated eIF4E is Mcl-1, which is preferentially translated in some cancer cells with defects in RAS/PI3K signaling (Rajasekhar et al. 2003; Adams and Cooper 2007) and whose translation is reduced in human leukemia cells following pharmacologic inhibition of MAPK signaling (Rahmani et al. 2005). Accordingly, MNK1-expressing tumors expressed elevated levels of eIF4E phosphorylation and Mcl-1 protein (Fig. 4A). Acute expression of eIF4E and MNK1, but not eIF4E S209A, in MEFs also increased Mcl-1 protein levels (Fig. 4B), whereas the dominant-negative MNK1 (T2A2) had the opposite effect (Fig. 4C). The ability of eIF4E or MNK1 to increase Mcl-1 protein was partly due to translation, since cells expressing these genes contain more Mcl-1 mRNA in the polysome fraction compared with controls, which remained unchanged in total mRNA (Fig. 4D,E). Finally, MCL-1 dramatically accelerated lymphomagenesis in the Eμ-Myc model (Fig. 4F). While MCL-1 expression in the resulting tumors was significantly higher than those expressing eIF4E or Mnk1 (data not shown), these data collectively indicate that Mcl-1 is one target of phosphorylated eIF4E during oncogenesis.
Figure 4.
Mcl-1 contributes to the oncogenic effect of eIF4E phosphorylation. (A) Immunoblotting of control and Mnk1 (Mnk1/T332D)-expressing lymphomas. (B) Immunoblotting of MEFs transduced with vector, eIF4E (eIF4E), the S209a mutant of eIF4E (S209A), and Mnk1 (Mnk1/T332D), or Mnk1/2 DKO MEFs. (C) Immunoblotting of 3T3 fibroblasts transduced with vector, dominant-negative Mnk1 (Mnk1/T2A2) or active Mnk1 (T332D) with (+) or without (−) serum. (D) Semiquantitative RT–PCR of Mcl-1 and β-Actin (Actin) mRNA in total RNA or polysomal RNA collected from MEFs transduced with vector, eIF4E (eIF4E), or Mnk1 (Mnk1/T332D); dilutions (1:1 to 1:100) were to enhance quantification. (E) Quantitative real-time RT–PCR of Mcl-1 normalized to β-Actin from cDNA samples prepared from MEFs transduced with vector, eIF4E, or active Mnk1 (Mnk1/T332D). Shown is a “relative quantification” whereby the Mcl-1 expression in triplicates of the total and polysome samples from vector-transduced cells is set to “1” and compared with the total and polysome fraction from cells expressing either eIF4E or Mnk1 (T332D). (F) Kaplan-Meier analysis of tumor latency of mice reconstituted with Eμ-Myc HSCs transduced with vector (black, n = 61, as in Fig. 1) or Mcl-1 (red, n = 5).
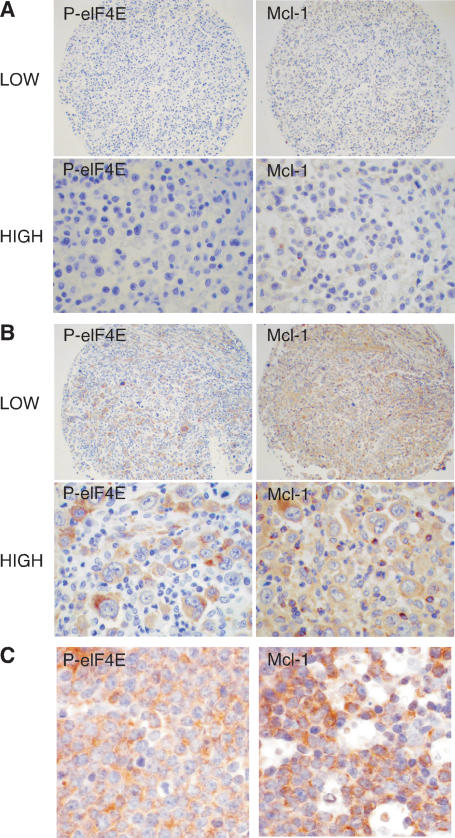
To determine whether eIF4E phosphorylation correlates with MCL-1 in human lymphomas, we analyzed tissue microarrays (TMAs) constructed from 77 patient samples of diffuse large B-cell lymphoma (DLBCL) (Fig. 5A,B). Notably, all 13 cases that were positive for phospho-eIF4E stained positive for MCL-1 protein and, among the 55 cases that were negative for eIF4E phosphorylation, 18 were negative and 37 were positive for MCL-1. Thus, eIF4E phosphorylation correlated with MCL-1 expression (P = 0.02 by Fisher’s exact test) in DLBCL, although other mechanisms must also contribute to the elevated levels of MCL-1 observed in this tumor type. Similarly, in a small cohort of Burkitt lymphoma samples (n = 8), all six cases that stained positive for phosphorylated eIF4E were also positive for MCL-1 compared with reactive lymph nodes (Fig. 5C). These clinical data are consistent with the genetic interactions observed in the mouse.
Figure 5.
eIF4E phosphorylation correlates with MCL-1 expression in human lymphoma. (A,B) Representative photographs (low and high power) of tissue arrays constructed from human DLBCL and probed for phosphorylated eIF4E and MCL-1. The photographs illustrate DLBCL staining either negative (A) or positive (B) for both phosphorylated eIF4E and MCL1. (C) Photomicrographs of Burkitt lymphoma samples stained for phosphorylated eIF4E and MCL-1.
These results provide insights into how translational deregulation promotes tumorigenesis and have therapeutic implications. For example, the biological roles and mechanism of action of eIF4E phosphorylation on Ser 209 have been controversial, leading to speculation that this event is not important for translational control in mammalian systems (Waskiewicz et al. 1997; Scheper and Proud 2002). However, while eIF4E phosphorylation is dispensable for mouse development (Ueda et al. 2004), our studies indicate that it plays a crucial role during tumorigenesis. Thus, eIF4E’s oncogenic activity correlates with its ability to activate translation and become phosphorylated on Ser 20, and constitutively activated Mnk1, a Ser 209 kinase, promotes tumorigenesis in a manner similar to eIF4E. Furthermore, a dominant-negative MNK1 mutant prevents eIF4E phosphorylation and inhibits the proliferation of tumor cells driven by mutations that deregulate translation. Consistent with our findings, the S209A cannot transform NIH 3T3 cells in vitro (Topisirovic et al. 2004). Still, it is unlikely that the organismal role of eIF4E phosphorylation is to promote tumorigenesis, but perhaps instead acts to enhance translation under circumstances of proliferation or cell stress.
In the Eμ-Myc model, phosphorylated eIF4E appears to promote tumorigenesis mainly by suppressing apoptosis, although how this occurs at the biochemical level remains to be fully elucidated. Most, but not all, studies suggest that phosphorylation has a positive effect on cap-dependent translation (Scheper and Proud 2002). Mcl-1, which is encoded by a message containing a structured 5′UTR, is specifically affected by the state of eIF4E phosphorylation in our model. Accordingly, high levels of phosphorylated eIF4E and MCL-1 occur together in human lymphomas. Thus, while MCL-1 may be regulated at additional levels (Le Gouill et al. 2004), its enhanced translation likely contributes to eIF4E-mediated oncogenesis.
The roles of the MNK kinases have remained elusive owing to the lack of biological readouts for their activity and a phenotype in the knockout mice (Ueda et al. 2004). We see that a constitutively activated MNK1 can be a potent oncogene in vivo. Of note, the similar onset and pathology of Mnk1- and eIF4E-expressing tumors, the inability of the MNK target site eIF4E mutant (S209A) to be oncogenic, and the shared ability of eIF4E and activated MNK1 to up-regulate MCL-1 all suggest that the oncogenic impact of MNK kinases involves targeting eIF4E, although it remains possible that nontranslational targets also play a role. Although sporadic mutations in Mnk1 or Mnk2 have not been observed, our studies raise the possibility these enzymes may mediate oncogenic signals in human cancers.
The fact that eIF4E acts at a convergence point of two crucial signaling pathways altered in cancer cells makes it an intriguing therapeutic target. Although targeting the translation initiation apparatus might produce substantial toxicity, our observation that eIF4E phosphorylation is important for tumorigenesis and previous work suggesting that it is dispensable for organismal viability suggests that inhibiting this event would have selective anti-tumor activity. Consistent with this idea and extending it further, our limited analysis to date suggests that the anti-proliferative effects of MNK1 kinase inhibition are restricted to tumors with deregulated translational control. Interestingly, small molecule inhibitors of the MNK kinases have been produced but are not being developed and, in our hands, were unable to prevent MNK-induced phosphorylation of eIF4E in vivo (A. Malina and J. Pelletier, unpubl.). By contrast, inhibitors of mTOR are in clinical trials, although their activity is limited, in part, by activation of AKT/PKB through a feedback mechanism involving S6 kinase (Sun et al. 2005). Targeting eIF4E downstream from mTOR—for example, via inhibition of MNK kinases—might circumvent feedback activation and present a therapeutic alternative.
Materials and methods
Animal studies
Lymphomas were generated and analyzed as described (Schmitt et al. 2002). The retroviruses used to produce tumors were based on MSCV-IRES-GFP (MIG) and expressed cDNAs encoding the eIF4E mutants W56A, W73A, S209A, and S209D, and the Mnk alleles T332D and T2A2. p53 loss of heterozygosity (LOH) was determined by allele-specific PCR of FACS-purified tumor cells (Wendel et al. 2004). For competition experiments, short-term lymphoma cultures were transduced with GFP-expressing retroviral vectors and the mixed populations were transplanted by tail-vein injection into nontransgenic female C57BL/6 animals. Tumors were then harvested and GFP expression was determined by flow cytometry (Wendel et al. 2004).
Histopathology
Preparation of samples, H&E stain, and detection of antibody stains was described (Wendel et al. 2004). Antibodies were against phosphorylated ribosomal S6 protein (Cell Signaling #2215) and phosphorylated eIF4E (Cell Signaling #9714). TUNEL was performed using a published protocol (McCurrach and Lowe 2001).
TMA analysis
The study cohort comprised 77 DLBCL collected and diagnosed at first evaluation at MSKCC and approved by the MSKCC IRB. TMAs were constructed as described previously (Maeda et al. 2005) and stained with antibodies against phosphorylated eIF4E (Cell Signaling #9741, 1:10) and Mcl1 (Cell Signaling #4572, 1:250). Positivity was defined as at least 30% of tumor cells stained for the probed antigen.
Western blot analysis
Immunoblots were performed as described (Wendel et al. 2004) using antibodies against phosphorylated Mnk1 (Cell Signaling #2111, 1:1000), total and phosphorylated eIF4E (both 1:1000, Cell Signaling #9742 and #9741), total and phosphorylated ribosomal S6 (Cell Signaling #2215, 1:1000), Mcl-1 (Abcam, ab32087-100, 1:1000), Tubulin (Sigma, B-5-1-2, 1:5000), and anti-HA (Roche 12013819, 1:5000).
Flow cytometry
Tumor cell suspensions were stained with the indicated antibodies (PharMingen and CalTag) conjugated with phycoerythrin (PE), Tri-Color (TC), or biotin, developed with streptavidin–allophycocyanin (PharMingen) and analyzed with FACSCalibur (Becton Dickinson).
Cell culture and viability
Primary MEFs of wild-type and Mnk1/2 DKO were cultured and analyzed as described (McCurrach and Lowe 2001). For apoptosis assays, MEFs were transduced with pBabe-Myc, selected with puromcin, and subjected to serum withdrawal for 12–24 h; viability was determined by trypan blue stain.
Polysome fractionation
As described in detail in Xi et al. (2004), cells were treated with cycloheximide before harvesting and lysing. The nuclei were pelleted and the ribosomal components were separated from the soluble fraction by ultracentrifugation. RNA was isolated using Trizol (Invitrogen) and cDNA synthesis was performed using the SuperScript III kit (Invitrogen). RT–PCR primers were Mcl1F, AGCCTGACTTCCCAGCTCAC; Mcl1R, GCACTCAGACCACATGCTTC; ActinF, AAGCTAACCGGGAGAA GATG; and ActinR, GTAGTCAGTGAGGTCGCGAC.
Acknowledgments
We thank N. Sonenberg for cDNAs, and K. Diggins, J. Simon, Miao-Miao, L. Bianco, and the CSHL animal facility. We also thank members of the Lowe and Wendel laboratories for helpful discussions and editorial advice. This work was supported by a Special Fellowship from the Leukemia and Lymphoma Society (H.G.W.), funds from the Special Projects Committee of Memorial Sloan-Kettering (H.G.W.), a CHIR grant (MOP-79385) (J.P.), the Don Monti Research Foundation (S.W.L.), and program project grants CA13106 and CA87497 from the National Cancer Institute (S.W.L). S.W.L. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1604407
References
- Adams K.W., Cooper G.M., Cooper G.M. Rapid turnover of mcl-1 couples translation to cell survival and apoptosis. J. Biol. Chem. 2007;282:6192–6200. doi: 10.1074/jbc.M610643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilanges B., Stokoe D., Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007;26:5973–5990. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- Dilling M.B., Dias P., Shapiro D.N., Germain G.S., Johnson R.K., Houghton P.J., Dias P., Shapiro D.N., Germain G.S., Johnson R.K., Houghton P.J., Shapiro D.N., Germain G.S., Johnson R.K., Houghton P.J., Germain G.S., Johnson R.K., Houghton P.J., Johnson R.K., Houghton P.J., Houghton P.J. Rapamycin selectively inhibits the growth of childhood rhabdomyosarcoma cells through inhibition of signaling via the type I insulin-like growth factor receptor. Cancer Res. 1994;54:903–907. [PubMed] [Google Scholar]
- Evan G.I., Wyllie A.H., Gilbert C.S., Littlewood T.D., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C., Wyllie A.H., Gilbert C.S., Littlewood T.D., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C., Gilbert C.S., Littlewood T.D., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C., Littlewood T.D., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C., Brooks M., Waters C.M., Penn L.Z., Hancock D.C., Waters C.M., Penn L.Z., Hancock D.C., Penn L.Z., Hancock D.C., Hancock D.C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N., Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Lachance P.E., Miron M., Raught B., Sonenberg N., Lasko P., Miron M., Raught B., Sonenberg N., Lasko P., Raught B., Sonenberg N., Lasko P., Sonenberg N., Lasko P., Lasko P. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 2002;22:1656–1663. doi: 10.1128/MCB.22.6.1656-1663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine K.S., Sonenberg N., Montine K.S., Sonenberg N., Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Le Gouill S., Podar K., Harousseau J.L., Anderson K.C., Podar K., Harousseau J.L., Anderson K.C., Harousseau J.L., Anderson K.C., Anderson K.C. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle. 2004;3:1259–1262. doi: 10.4161/cc.3.10.1196. [DOI] [PubMed] [Google Scholar]
- Maeda T., Hobbs R.M., Merghoub T., Guernah I., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P.P., Hobbs R.M., Merghoub T., Guernah I., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P.P., Merghoub T., Guernah I., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P.P., Guernah I., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P.P., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P.P., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P.P., Teruya-Feldstein J., Pandolfi P.P., Pandolfi P.P. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N., Petroulakis E., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N., Larsson O., Rajasekhar V.K., Sonenberg N., Rajasekhar V.K., Sonenberg N., Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K., Gingras A.C., Sonenberg N., Burley S.K., Sonenberg N., Burley S.K., Burley S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- McCurrach M.E., Lowe S.W., Lowe S.W. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 2001;66:197–227. doi: 10.1016/s0091-679x(01)66010-2. [DOI] [PubMed] [Google Scholar]
- Murata T., Shimotohno K., Shimotohno K. Ubiquitination and proteasome-dependent degradation of human eukaryotic translation initiation factor 4E. J. Biol. Chem. 2006;281:20788–20800. doi: 10.1074/jbc.M600563200. [DOI] [PubMed] [Google Scholar]
- Polunovsky V.A., Gingras A.C., Sonenberg N., Peterson M., Tan A., Rubins J.B., Manivel J.C., Bitterman P.B., Gingras A.C., Sonenberg N., Peterson M., Tan A., Rubins J.B., Manivel J.C., Bitterman P.B., Sonenberg N., Peterson M., Tan A., Rubins J.B., Manivel J.C., Bitterman P.B., Peterson M., Tan A., Rubins J.B., Manivel J.C., Bitterman P.B., Tan A., Rubins J.B., Manivel J.C., Bitterman P.B., Rubins J.B., Manivel J.C., Bitterman P.B., Manivel J.C., Bitterman P.B., Bitterman P.B. Translational control of the antiapoptotic function of Ras. J. Biol. Chem. 2000;275:24776–24780. doi: 10.1074/jbc.M001938200. [DOI] [PubMed] [Google Scholar]
- Rahmani M., Davis E.M., Bauer C., Dent P., Grant S., Davis E.M., Bauer C., Dent P., Grant S., Bauer C., Dent P., Grant S., Dent P., Grant S., Grant S. Apoptosis induced by the kinase inhibitor BAY 43-9006 in human leukemia cells involves down-regulation of Mcl-1 through inhibition of translation. J. Biol. Chem. 2005;280:35217–35227. doi: 10.1074/jbc.M506551200. [DOI] [PubMed] [Google Scholar]
- Rajasekhar V.K., Viale A., Socci N.D., Wiedmann M., Hu X., Holland E.C., Viale A., Socci N.D., Wiedmann M., Hu X., Holland E.C., Socci N.D., Wiedmann M., Hu X., Holland E.C., Wiedmann M., Hu X., Holland E.C., Hu X., Holland E.C., Holland E.C. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Pandolfi P.P., Pandolfi P.P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Londei P., Cordon-Cardo C., Pandolfi P.P., Cordon-Cardo C., Pandolfi P.P., Pandolfi P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Scheper G.C., Proud C.G., Proud C.G. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 2002;269:5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C.A., Fridman J.S., Yang M., Baranov E., Hoffman R.M., Lowe S.W., Fridman J.S., Yang M., Baranov E., Hoffman R.M., Lowe S.W., Yang M., Baranov E., Hoffman R.M., Lowe S.W., Baranov E., Hoffman R.M., Lowe S.W., Hoffman R.M., Lowe S.W., Lowe S.W. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Bath M.L., Cory S., Harris A.W., Bath M.L., Cory S., Bath M.L., Cory S., Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- Sun S.Y., Rosenberg L.M., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R., Rosenberg L.M., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R., Zhou Z., Yue P., Fu H., Khuri F.R., Yue P., Fu H., Khuri F.R., Fu H., Khuri F.R., Khuri F.R. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- Topisirovic I., Ruiz-Gutierrez M., Borden K.L., Ruiz-Gutierrez M., Borden K.L., Borden K.L. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64:8639–8642. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R., Fukuyama H., Nagata S., Fukunaga R., Nagata S., Fukunaga R., Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Flynn A., Proud C.G., Cooper J.A., Flynn A., Proud C.G., Cooper J.A., Proud C.G., Cooper J.A., Cooper J.A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Johnson J.C., Penn B., Mahalingam M., Kimball S.R., Cooper J.A., Johnson J.C., Penn B., Mahalingam M., Kimball S.R., Cooper J.A., Penn B., Mahalingam M., Kimball S.R., Cooper J.A., Mahalingam M., Kimball S.R., Cooper J.A., Kimball S.R., Cooper J.A., Cooper J.A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel H.G., De Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., De Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Cordon-Cardo C., Pelletier J., Lowe S.W., Pelletier J., Lowe S.W., Lowe S.W. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wendel H.G., Malina A., Zhao Z., Zender L., Kogan S.C., Cordon-Cardo C., Pelletier J., Lowe S.W., Malina A., Zhao Z., Zender L., Kogan S.C., Cordon-Cardo C., Pelletier J., Lowe S.W., Zhao Z., Zender L., Kogan S.C., Cordon-Cardo C., Pelletier J., Lowe S.W., Zender L., Kogan S.C., Cordon-Cardo C., Pelletier J., Lowe S.W., Kogan S.C., Cordon-Cardo C., Pelletier J., Lowe S.W., Cordon-Cardo C., Pelletier J., Lowe S.W., Pelletier J., Lowe S.W., Lowe S.W. Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo. Cancer Res. 2006;66:7639–7646. doi: 10.1158/0008-5472.CAN-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q., Cuesta R., Schneider R.J., Cuesta R., Schneider R.J., Schneider R.J. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes & Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]