Abstract
Retrosequences generated by reverse transcription of mRNA transcripts have a substantial influence on gene expression patterns, generation of novel gene functions, and genome organization. The Ty1 retrotransposon is a major source of RT activity in the yeast, Saccharomyces cerevisiae, and Ty1 retromobility is greatly elevated in strains lacking telomerase. We report that Ty1-dependent formation of retrosequences derived from single-copy gene transcripts is progressively elevated as yeast cells senesce in the absence of telomerase. Retrosequences are frequently fused to Ty1 sequences, and occasionally to sequences from other mRNA transcripts, forming chimeric pseudogenes. Efficient retrosequence formation requires the homologous recombination gene RAD52. Selection for retrosequence formation is correlated with a high frequency of chromosome rearrangements in telomerase-negative yeast. Ty1-associated retrosequences were present at the breakpoint junctions of four chromosomes analyzed in detail. Our results support a role for reverse transcripts in promoting chromosome rearrangements.
[Keywords: Retrosequence, pseudogene, Ty1, telomeres, Saccharomyces cerevisiae]
Reverse transcription of cellular mRNA and subsequent incorporation of the resulting complementary DNA (cDNA) into chromosomal DNA generates retrosequences, which comprise functional retrogenes as well as processed (or retrotransposed) pseudogenes, with the latter being defined by their inability to encode functional proteins. Approximately 200, 1200, 5000, and 7800 processed pseudogenes have been reported in the Caenorhabditis elegans, rice, mouse, and human genomes, respectively (Harrison et al. 2001; Ohshima et al. 2003; Torrents et al. 2003; Zhang et al. 2003, 2004; Wang et al. 2006). Retrosequences derived from mRNA sequences typically lack regulatory sequences and introns, and contain oligo(A/T) tracts at their 3′ ends. Retrosequences formed by the RT activity of non-long-terminal repeat (LTR) retrotransposons are usually flanked by target site duplications (Esnault et al. 2000), while those formed by LTR retrotransposons or retroviruses are typically flanked by sequences of the retroelements themselves (Derr et al. 1991; Jin and Bennetzen 1994; Wang et al. 2006).
Most of the studies that contributed to our understanding of the process and consequences of retrosequence formation are retrospective. Experimental systems to detect retrosequence formation are limited by extremely low frequencies of events (Schacherer et al. 2004), or they rely on overexpression of the donor genes and/or retroelements (Derr et al. 1991; Esnault et al. 2000). We reported that retrosequences of subtelomeric Y′ elements form frequently in Saccharomyces cerevisiae strains that lack telomerase (Maxwell et al. 2004). Formation of Y′ and other mRNA-derived retrosequences in yeast depends on the yeast Ty1 LTR retrotransposon as a source of RT activity and the primer for reverse transcription (Derr et al. 1991; Maxwell et al. 2004).
Following telomerase inactivation, S. cerevisiae strains become senescent as telomeric DNA erodes, but after 50–100 generations, a fraction of cells, termed survivors, escape growth arrest and maintain altered telomere structures by homologous recombination (Lundblad and Blackburn 1993; Teng and Zakian 1999). Ty1 mRNA is reverse-transcribed into cDNA in cytoplasmic virus-like particles, and both Ty1 cDNA synthesis and retromobility progressively increase as telomerase-negative S. cerevisiae strains senesce (Scholes et al. 2003). Telomerase-negative yeast strains also have elevated levels of chromosome rearrangements during late senescence (Hackett et al. 2001). Ty1 sequences are frequently present at the junctions of chromosome rearrangements obtained by genetic selection systems or by selection for adaptation to specific growth conditions (Dunham et al. 2002; Umezu et al. 2002; Lemoine et al. 2005; Putnam et al. 2005). Therefore, we hypothesized that Ty1 RT activity promotes chromosome rearrangements in telomerase-negative yeast. We asked whether the mechanism of retrosequence formation would promote chromosome rearrangements in telomerase-negative yeast, since multiple pathways of retrosequence insertion into the genome, including recombination, have been proposed (Derr et al. 1991; Maxwell et al. 2004). To address this question, we analyzed retrosequence formation from donor genes unrelated to telomere maintenance during senescence and survivor formation.
Here, we demonstrate that Ty1-dependent formation of retrosequences derived from transcripts of single-copy genes occurs at progressively higher frequencies as yeast cells senesce in the absence of telomerase. Retrosequence formation is correlated with the appearance of chromosome rearrangements in telomerase-negative yeast survivors. Genetic analyses and molecular characterizations of select chromosome rearrangements are consistent with a role for recombination between chimeric Ty1 retrosequence cDNA molecules and genomic Ty1 sequences in the production of these chromosome rearrangements.
Results
Elevated levels of retrosequence cDNA derived from single-copy genes in telomerase-negative yeast
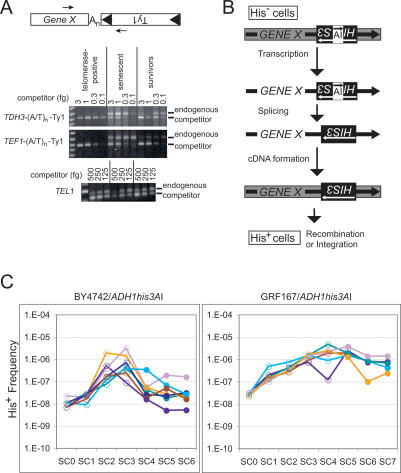
We asked whether reverse transcription of mRNA transcripts unrelated to telomeres and telomere maintenance was increased in telomerase-negative yeast. Telomerase-negative derivatives of isogenic strains were obtained by segregating a plasmid copy of TLC1 (see Materials and Methods), which encodes the RNA subunit of telomerase, and were subcultured by serial restreaking for single colonies. We used a PCR assay to detect retrosequence cDNA that is reverse transcribed by priming from Ty1 cDNA onto the poly(A) tail of an mRNA and continuing into the ORF (Fig. 1A), because Y′–Ty1 cDNA junctions frequently have this structure (Maxwell et al. 2004). We used primers for two genes identified as highly expressed genes, TDH3 (glyceraldehyde-3-phosphate dehydrogenase) and TEF1 (translation elongation factor EF-1α), and two genes not identified as highly expressed, FUS3 (a mitogen-activated protein kinase) and SUC2 (invertase) (Velculescu et al. 1997). We obtained PCR products for TEF1–Ty1 and TDH3–Ty1 cDNA junctions (Fig. 1A) and sequenced the products from two independent reactions for each gene. The products consisted of sequences from the 3′ end of each gene followed by an oligo(A/T) tract corresponding to the mRNA poly(A) tail adjacent to Ty1 LTR sequences. We could not detect PCR products for FUS3–Ty1 or SUC2–Ty1 cDNA junctions (data not shown), which may be because FUS3 and SUC2 mRNA levels are lower than TDH3 and TEF1 mRNA levels. Indeed, most human processed pseudogenes are produced from highly transcribed genes (Zhang et al. 2003).
Figure 1.
Retrosequence formation becomes more frequent as telomerase-negative strains senesce. (A) Arrows in the diagram indicate the orientation of primers used to detect retrosequence cDNA junctions between sequences from the 3′ UTR of hypothetical GENE X and the 3′ end of Ty1, spanned by an oligo(A) tract, An, corresponding to the poly(A) tail of the GENE X transcript. Panels below the diagram show results of competitive PCR using equivalent amounts of genomic DNA from telomerase-positive, telomerase-negative senescent, and telomerase-negative survivor populations (BY4742 background) using primers and competitors for the indicated cDNA junctions, or with primers and competitor for the TEL1 gene (shown in bottom panel). (B) The his3AI indicator gene consists of a HIS3 gene interrupted by an artificial intron (AI) in the opposite transcriptional orientation. The his3AI gene is placed into the 3′ UTR of a gene in an orientation that allows AI to be spliced from the gene’s transcript. Formation of a functional copy of HIS3 by reverse transcription of the spliced transcript and incorporation of the cDNA into the genome produces cells that are His+ prototrophs. (C) Semi-log graphs of the frequency of His+ prototroph formation in telomerase-positive (SC0) and serially subcultured telomerase-negative derivatives (SC1–7) of strains from two different genetic backgrounds (BY4742 and GRF167) containing ADH1his3AI are shown. Open circles represent telomerase-positive and senescent populations, while filled circles represent the subculture when survivors first appeared and all subcultures thereafter, as determined by Southern analysis of telomeres of each culture at each time point (data not shown).
The copy number of the TEF1–oligo(A/T)–Ty1 and TDH3–oligo(A/T)–Ty1 cDNA junctions (Fig. 1A) per genome was determined by a competitive PCR assay, using the single-copy TEL1 gene as a normalization control (Maxwell et al. 2004). The mean copies per genome of TDH3–oligo(A/T)–Ty1 and TEF1–oligo(A/T)–Ty1 cDNA junctions were 6.1 ± 1.5 × 10−5 and 7.7 ± 1.5 × 10−5 in three telomerase-positive strains, 3.1 ± 1.0 × 10−3 and 2.1 ± 0.5 × 10−3 in four telomerase-negative senescent populations, and 4.4 ± 1.7 × 10−4 and 5.7 ± 2.2 × 10−4 in four telomerase-negative survivors, respectively. The increase in copy number of TDH3–oligo(A/T)–Ty1 or TEF1–oligo(A/T)–Ty1 was ∼50-fold or 28-fold in senescent populations and approximately sevenfold in survivors, respectively. We conclude that reverse transcription of not only mRNA of Y′ elements, but also mRNA of single-copy genes, is induced in telomerase-negative yeast (Maxwell et al. 2004).
Progressive induction of retrosequence formation during senescence
We used a genetic assay to measure retrosequence formation during senescence of telomerase-negative yeast. Retrosequence formation, defined as the incorporation of retrosequence cDNA into the genome, was measured by inserting the his3AI retrotranscript indicator gene (Curcio and Garfinkel 1991) into the 3′ untranslated region (UTR) of the endogenous ADH1 (alcohol dehydrogenase 1) locus. This allowed us to measure the frequency of ADH1HIS3 retrosequence formation as the frequency of obtaining His+ prototrophs (Fig. 1B). ADH1 was chosen because it is a well-expressed gene (Velculescu et al. 1997) whose function is unrelated to telomere maintenance, and it is >100 kb from a telomere. Telomerase was inactivated in strains harboring ADH1his3AI by segregating a plasmid copy of TLC1 (BY4742 background), or by selecting for loss of the EST2 allele, which encodes the catalytic protein subunit of telomerase, from an EST2–URA3–est2Δ1 construct integrated into the genome at the EST2 locus (GRF167 background). Telomerase-positive populations are referred to as subculture 0 (SC0), and colonies screened for loss of TLC1 or EST2 are referred to as SC1. Six or seven telomerase-negative SC1 derivatives in each background were serially restreaked until survivors appeared. Once survivors appeared, the survivors were serially restreaked until a total of six or seven subcultures was obtained. The frequency of His+ prototroph formation for each population at each subculture was determined by using colonies to inoculate triplicate cultures and comparing the number of His+ prototrophs with the number of viable cells (Fig. 1C).
The His+ frequencies in both strain backgrounds progressively increased during senescence (Fig. 1C, open circles), though it was formally possible that inactivating different telomerase components (TLC1 or EST2) in each background could have yielded different phenotypes. For the BY4742 strains, His+ frequencies peaked after two or three subcultures and then noticeably declined (Fig. 1C, left panel). For the GRF167 strains, the highest His+ frequencies were obtained after four to five subcultures, but they then decreased only modestly (Fig. 1C, right panel). The median peak His+ frequencies for the BY4742-derived and GRF167-derived strains with ADH1his3AI were 5.2 × 10−7 and 2.4 × 10−6, respectively. These values represent 47-fold and 83-fold inductions in His+ frequencies, respectively.
Survivors appeared at SC4 in the BY4742 background and at SC5, SC6, or SC7 in the GRF167 background (Fig. 1C, survivors indicated by filled circles), as determined by probing Southern blots of genomic DNA for telomeric sequences (data not shown). The highest His+ frequency was obtained one or two subcultures prior to survivor formation for 12 of the 13 telomerase-negative populations analyzed (Fig. 1C). Ty1 retrotransposition exhibits a similar increase during senescence and subsequent decrease following survivor formation in the BY4742 background (Scholes et al. 2003). The similarity in the trends for Ty1 retrotransposition and retrosequence formation is consistent with Ty1’s role in retrosequence formation.
Frequencies of retrosequence formation are reduced in rad52 mutants
Previous work has indicated that gene conversion of a chromosomal allele by retrosequence cDNA requires RAD52 and RAD1 (Derr and Strathern 1993; Derr 1998). In addition, single-gene duplications resulting from retrosequence formation have been reported to require RAD52 (Schacherer et al. 2005). We tested whether RAD52-dependent homologous recombination would be required for the elevated frequency of ADH1HIS3 cDNA incorporation in telomerase-negative cells. Since RAD52 is required for survival of telomerase-negative strains (Teng and Zakian 1999), we analyzed rad52Δ telomerase-negative strains at the earliest time point during senescence. Deletion of RAD52 reduced the frequency of forming His+ prototrophs sevenfold or 10-fold in telomerase-positive strains and at least fivefold or 40-fold in telomerase-negative strains, depending on the strain background (Table 1). This effect is particularly striking in light of the observation that deletion of RAD52 activates Ty1 mobility (Rattray et al. 2000). In contrast, deletion of RAD1 did not alter the His+ frequencies in strains harboring ADH1his3AI (Table 1). Therefore, processing of the cDNA or recombination intermediates by the Rad1–Rad10 endonuclease complex does not play a role in ADH1 retrosequence formation.
Table 1.
Influence of DNA repair/recombination proteins on retrosequence formation
aTLC1 indicates the BY4742 strain background, and EST2 indicates the GRF167 strain background. Two or more independent transformants were analyzed for each mutation.
bSets of triplicate cultures tested.
cWhen no His+ prototrophs were obtained, the frequency was listed as less than the value that would have been obtained if one His+ prototroph had formed. Such frequency values were ranked as if they were equal to their maximum frequency value for determining the range and median.
dNormalized to a wild-type telomerase-positive strain, a tlc1Δ strain, or an est2Δ1 strain, as appropriate.
(*) P < 0.025; (**) P < 0.01; (***) P < 0.001.
We also analyzed retrosequence formation in strains harboring deletions of RAD51, MRE11, or DNL4. These genes have roles in homologous recombination, both homologous recombination and nonhomologous end joining, or nonhomologous end joining, respectively (Krogh and Symington 2004). In telomerase-positive strains, none of these three genes was required for retrosequence formation, and moderate increases in His+ frequencies were observed in telomerase-positive rad51Δ or mre11Δ strains (Table 1), consistent with activation of Ty1 mobility in these mutants (Rattray et al. 2000; Scholes et al. 2001). Deletion of RAD51 did not alter the frequency of retrosequence formation in telomerase-negative strains (SC2), and deletion of DNL4 increased the levels of retrosequence formation in senescent cells (Table 1). Therefore, retrosequence formation does not depend on nonhomologous end joining and does not require RAD51-dependent homologous recombination.
We confirmed that activation or inhibition of Ty1 mobility produced corresponding changes in the frequency of forming ADH1HIS3 retrosequences. Expression of a Ty1kanAI element under the control of a galactose-inducible promoter from a high-copy plasmid (pGAL1Ty1kanAI) in telomerase-positive strains harboring ADH1his3AI resulted in high levels of Ty1kanAI mobility (data not shown) and substantially elevated the His+ frequency (Supplementary Table S1). Activation of endogenous Ty1 element mobility by deletion of MED1 or RTT101 (Scholes et al. 2001) also elevated the frequency of ADH1HIS3 retrosequence formation (Supplementary Table S1). We were unable to obtain any His+ prototrophs from telomerase-negative senescent strains that harbored an spt3−101 allele (Supplementary Table S1), which prevents expression of endogenous Ty1 and Ty2 elements (Boeke et al. 1986), confirming that retrosequence formation depends on Ty1 and possibly Ty2 expression.
Selection for retrosequences is correlated with chromosome rearrangements in telomerase-negative survivors
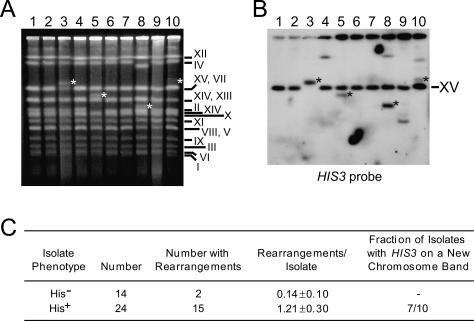
We reasoned that chimeric Ty1 retrosequence cDNA molecules might mediate individual recombination events involving multiple genomic sites, and thereby lead to chromosome deletions or translocations in some instances. We grew cultures of three independent His− telomerase-negative survivors derived from BY4742/ADH1his3AI strains to obtain His+ clones that harbored retrosequences and control His− clones that lacked retrosequences. To find evidence of chromosome rearrangements, we then used clamped homogeneous electric fields (CHEF) gels to compare intact chromosome-banding patterns from a total of 14 His− clones and 24 His+ clones with the banding pattern from each of the three original survivors. His+ clones harboring ADH1HIS3 retrosequences had an eightfold increase in the mean number of additional chromosome bands and missing chromosome bands compared with His− clones (Fig. 2). This increase in chromosome rearrangements is specific to the presence of retrosequences, rather than simply resulting from genome instability in the absence of telomerase, because multiple preparations from individual survivors yielded consistent chromosome-banding patterns. Moreover, chromosome rearrangements were rarely observed in His− clones that lacked retrosequences (Fig. 2). By Southern analysis, we detected HIS3 sequences on a new chromosome band in 70% of His+ isolates with one or more new chromosome bands (Fig. 2). These results are consistent with the hypothesis that retrosequence formation promotes chromosome rearrangements.
Figure 2.
Selection for ADH1HIS3 retrosequence formation is correlated with the appearance of chromosome rearrangements. (A) Lanes 1–5 and 6–10 each show intact chromosomes from five clones obtained from an independent His− telomerase-negative survivor separated using CHEF gel electrophoresis. Two clones in each set were obtained without selection for retrosequence formation (lanes 1,2,6,7) and therefore remained His−, while three clones in each set were His+ due to selection for retrosequences (lanes 3–5,8–10). Asterisks indicate new chromosome bands that also harbored HIS3 sequences (see below). The yeast chromosome or pair of chromosomes corresponding to each band is indicated to the right of the gel image. (B) A Southern blot of the gel shown in A was probed for HIS3 sequences. Both ADH1his3AI and a partial deletion allele of HIS3, his3Δ1, are present on chromosome XV, so the band corresponding to chromosome XV was detected in all lanes. (Lanes 3–5,8–10) At least one additional band was detected in each sample from His+ clones, with the exception of the sample in lane 3. The lack of an additional band in lane 3 resulted from the incorporation of ADH1HIS3 into chromosome XV. Asterisks indicate the same new chromosome bands indicated in A. (C) Chromosomal rearrangement frequencies in isolates obtained from a total of three independent His− telomerase-negative survivors. New bands or missing bands on CHEF gels were scored as rearrangements. The mean (±standard error) number of rearrangements per isolate is shown, and the difference in the means is statistically significant (P = 0.001).
Incorporation of retrosequences into the genome leads to frequent gene duplications in telomerase-negative strains
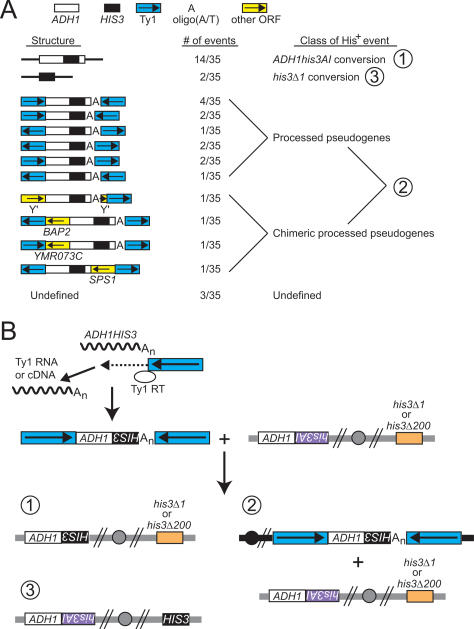
Based on the requirement for Rad52 in ADH1HIS3 retrosequence formation (Table 1) and on the fusion of Ty1 sequences to retrosequence cDNA (Fig. 1A), we expected ADH1HIS3–Ty1 cDNA to recombine with genomic ADH1his3AI, his3Δ1, or Ty1 sequences. Using a series of PCR reactions, we obtained sequences flanking both sides of the spliced HIS3 gene in 32 of 35 His+ derivatives of telomerase-negative survivors derived from BY4742/ADH1his3AI strains. Spliced HIS3 sequences replaced his3AI sequences at the ADH1 locus in 40% (14 of 35) of these His+ survivors (Fig. 3A). These events likely arose through use of the ADH1HIS3–Ty1 cDNA as a donor for gene conversion recombination with the ADH1his3AI allele (Fig. 3B). We refer to these events as ADH1his3AI conversions (Fig. 3A). Spliced HIS3 sequences were found at the endogenous HIS3 locus in 6% (two of 35) of His+ survivors (Fig. 3A). These events could have formed if the ADH1HIS3–Ty1 cDNA was used as a donor for gene conversion recombination with the his3Δ1 allele (Fig. 3B). We refer to these events as his3Δ1 conversions. Finally, spliced HIS3 sequences were flanked by Ty1 sequences or ADH1 sequences fused to Ty1 in 46% (16 of 35) of His+ survivors (Fig. 3A). Formation of these events was likely the result of recombination between ADH1HIS3–Ty1 cDNA and genomic Ty1 sequences (Fig. 3B). We refer to these events as processed pseudogenes because formation of the HIS3 retrosequence did not occur by replacement of genomic his3AI or his3Δ1 sequences (Fig. 3A).
Figure 3.
Retrosequences typically replace the donor allele or are flanked by Ty1 sequences. (A) The number of independent His+ events with particular flanking DNA sequences is illustrated. A key at the top indicates which sequences correspond to which symbols, and transcriptional orientations are indicated with arrows. Sequences to the left or right are upstream or downstream relative to ADH1HIS3, respectively. “Undefined” refers to events for which only the upstream or the downstream junction was characterized, and the numbers for different classes correspond to the numbers in B. (B) Ty1 RT is shown using Ty1 cDNA as a primer to initiate reverse transcription of the ADH1HIS3 mRNA starting at the poly(A) tail. At variable points during reverse transcription, Ty1 RT switches templates and begins copying Ty1 RNA or cDNA at sites of microhomology between ADH1HIS3 and Ty1 sequences. One example of the possible junction sites and relative orientations of the retrosequence cDNA and the Ty1 sequences is shown. The botom portion of the diagram is a representation of the recombination events that produced the structures shown in A: ADH1his3AI conversion (1), processed pseudogene formation (2), and his3Δ1 conversion (3). Note that the his3Δ1 allele is present in strains from the BY4742 background, but a complete HIS3 deletion (his3Δ200) is present in strains from the GRF167 background.
We characterized the structures of all 16 processed pseudogenes. Four of these proved to have exceptional sequence structures, and we refer to these four as chimeric processed pseudogenes (Fig. 3A). For the 12 simple processed pseudogenes, upstream junctions typically consisted of the ADH1 ORF or 5′ UTR joined to Ty1 sequences at sites where ADH1 and Ty1 sequences contained 2–7 base pairs (bp) of microhomology in a 10-bp window centered around the junction (Supplementary Fig. S1A; data not shown). In contrast, the downstream junctions consisted of either HIS3 promoter or ADH1 3′ UTR sequences followed by oligo(A/T) tracts joined to Ty1 sequences, with 0–3 bp of microhomology between the Ty1 sequences and the oligo(A/T) tract (Supplementary Fig. S1A; data not shown). One chimeric ADH1HIS3 processed pseudogene consisted of ADH1HIS3 sequences fused to transcribed sequences from a Y′ element at the upstream and downstream junctions, with Ty1 sequences adjacent to Y′ sequences at the downstream junction (Fig. 3A; Supplementary Fig. S1B). The other three chimeric pseudogenes had transcribed regions of different single-copy genes upstream of or downstream from ADH1HIS3 cDNA, which were then flanked by Ty1 sequences (Fig. 3A; Supplementary Fig. S1C,D; data not shown). The exceptional events provide evidence that Ty1-dependent retrosequence formation can generate chimeric gene duplications.
Eleven of 16 ADH1HIS3 processed pseudogenes were flanked by Ty1 sequences in opposite transcriptional orientations relative to each other (Fig. 3A). If insertion of a processed pseudogene occurred by recombination with one genomic Ty1 element or LTR, we would expect the processed pseudogene to be flanked by Ty1 sequences in direct orientation. The presence of flanking Ty1 sequences in opposite orientations is consistent with retrosequence formation that involved recombination with two (or more) genomic Ty1 sequences. Recombination events involving two genomic sites could produce chromosomal deletions and translocations, which is consistent with our identification of frequent chromosome rearrangements in cells harboring retrosequences (Fig. 2).
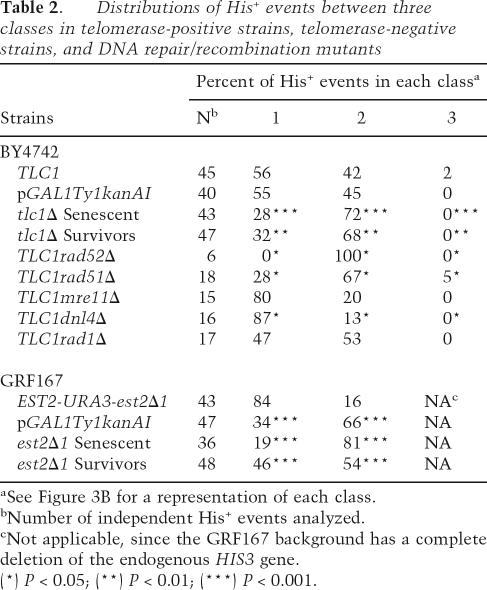
We analyzed additional independent His+ derivatives of telomerase-positive and telomerase-negative strains to determine the relative frequency of ADH1his3AI conversions, his3Δ1 conversions, and processed pseudogene events in each strain. The his3Δ1 conversions occurred only in the BY4742 background (since strains in the GRF167 background harbor a complete deletion of HIS3 [his3Δ200]) and were rarely observed (Table 2, class 3 columns). The fraction of processed pseudogenes relative to ADH1his3AI conversions was increased in senescent populations and survivors relative to telomerase-positive cells in both strain backgrounds (Table 2). Overexpression of a plasmid-borne, galactose-inducible Ty1kanAI element caused a similar increase in processed pseudogene events only in the GRF167 background (Table 2, class 2 column).
Table 2.
Distributions of His+ events between three classes in telomerase-positive strains, telomerase-negative strains, and DNA repair/recombination mutants
aSee Figure 3B for a representation of each class.
bNumber of independent His+ events analyzed.
cNot applicable, since the GRF167 background has a complete deletion of the endogenous HIS3 gene.
(*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
We also analyzed smaller sets of independent His+ events in BY4742-derived telomerase-positive strains with deletions of RAD52, RAD51, MRE11, DNL4, or RAD1. Processed pseudogene events were more common than ADH1his3AI conversion events in rad52Δ and rad51Δ strains, and the converse was seen in mre11Δ and dnl4Δ strains (although the difference was not statistically significant for mre11Δ strains). No substantial change in distribution was observed in the rad1Δ strains (Table 2). His+ events were rarely obtained in rad52Δ strains, and the processed pseudogene events identified could have resulted from rare instances of nonhomologous end joining or the activity of Ty1 integrase.
Retrosequences are present at chromosome rearrangement junctions
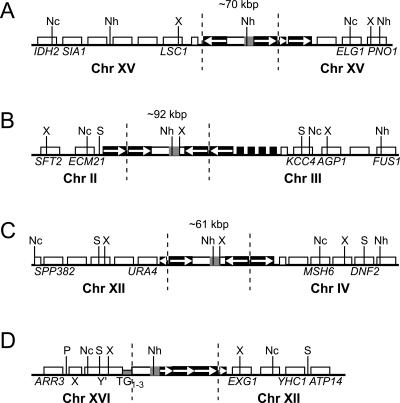
We performed detailed analyses of the four rearranged chromosomes indicated with asterisks in Figure 2 to determine if retrosequences were present at the breakpoint junctions. Analysis of missing chromosome bands on CHEF gels and a series of Southern blots of intact chromosomes were used to identify the chromosome or chromosomes of origin for each rearranged chromosome (Fig. 2; Supplementary Fig. S2A; data not shown). Approximate junction sites for each rearrangement were determined by additional Southern blots of intact chromosomes (Supplementary Fig. S2A; data not shown). We then obtained unique flanking sequences using a modified inverse PCR strategy. Genomic DNA from each His+ derivative was digested with a restriction enzyme that was not expected to digest ADH1HIS3 or Ty1 sequences, ligated to form circles, and amplified with one primer to HIS3 sequences and a second primer to an ORF expected to flank one side of the ADH1HIS3 insertion, and the resulting PCR products (Supplementary Fig. S2B) were verified by sequencing. Restriction mapping of the junctions by Southern blotting of CHEF gels using multiple enzymes that do not have sites in Ty1 (Supplementary Fig. S3) was consistent with the presence of many copies of Ty1 sequences flanking the ADH1HIS3 retrosequences, leading to total insertion sizes of >50 kb in three of four cases.
An ADH1HIS3 processed pseudogene spanned a breakpoint junction on each of the four chromosomes, and three of the four rearrangements clearly resulted from translocations (Fig. 4B–D). We identified contiguous chromosome XV sequences on both sides of ADH1HIS3 for chromosome A (Fig. 4A), so the slower migration of this chromosome evident in Figure 2 (lane 3) could have resulted from the insertion of a long stretch of Ty1 and ADH1HIS3 sequences at this site. Chromosome B (Fig. 4B) resulted from insertion of duplicated sequences from the left arm of chromosome III into the left arm of chromosome II. Only the chromosome II and III junction that is spanned by ADH1HIS3 is drawn. Chromosome C (Fig. 4C) resulted from attachment of duplicated sequences from the end of the right arm of chromosome XII to a site in the right arm of chromosome IV. Chromosome D (Fig. 4D) resulted from a fusion between duplicated sequences from the right arm of chromosome XII and the right arm telomere of chromosome XVI. The breakpoint of chromosome B was ∼30 kb from the chromosome II left-arm telomere, the breakpoint of chromosome C was ∼115 kb from the chromosome XII right-arm telomere (chromosome XII is >1000 kb), and one breakpoint of chromosome D was the right-arm telomere of chromosome XVI. Therefore, these rearrangements might have resulted from repair of breaks induced by destabilized telomeres in these telomerase-negative strains.
Figure 4.
ADH1HIS3 retrosequences are present at chromosome breakpoint junctions. Schematic representations of rearrangement junctions for the four chromosomes marked in Figure 2 are shown, as determined by Southern blotting of CHEF gels and inverse PCR. Chromosomes A, B, C, and D in the figure correspond to chromosomes with asterisks in lanes 3, 5, 8, and 10 of Figure 2, respectively. The chromosome of origin of sequences flanking each side of ADH1HIS3 is indicated below each drawing. Indicated sequence features correspond to sequences present in the corresponding regions of the sequenced S. cerevisiae genome (http://www.yeastgenome.org). Long, medium-length, and short white boxes represent a Y′ element, verified ORFs, and tRNA genes, respectively. Medium-length and short black boxes represent Ty1 or Ty2 elements and solo LTRs, respectively. A short gray box indicates telomeric repeats (TG1–3). ADH1HIS3 is indicated with a box containing a white region (ADH1) and a gray region (HIS3). White arrows indicate transcriptional orientations of select Ty1 sequences. Dotted vertical lines distinguish the sequences inserted through retrosequence formation from surrounding genomic sequences. Solid vertical lines indicate relevant restriction enzyme sites: NcoI (Nc), NheI (Nh), PvuII (P), StuI (S), and XbaI (X). For A, B, and C, the total number of Ty elements at the insertion site could not be determined, but the approximate size of each insertion in kilobase pairs is given, based on Southern blotting of restriction fragments separated on CHEF gels (see Supplementary Fig. S3). For the sake of simplicity, we show only a single Ty1 element flanking ADH1HIS3 in these three cases. Diagrams are not drawn to scale.
Solo LTRs or complete Ty1 or Ty2 elements are present in the sequenced S. cerevisiae genome at sites corresponding to the junction sites shown in Figure 4 (http://www.yeastgenome.org). The orientation of Ty1 sequences flanking the ADH1HIS3 sequences were consistent with the orientations of Ty sequences in the sequenced genome for chromosomes A, B, and D, but not for C (http://www.yeastgenome.org). For chromosome C, perhaps the processed pseudogene insertion arose through a more complicated recombination event involving arrays of Ty1 cDNAs flanking the processed pseudogene. Homologous recombination between Ty1 retrosequence cDNA and genomic Ty sequences could have produced all of these junctions, except for the left junction drawn for chromosome D; lack of a long stretch of homology between ADH1 sequences and telomeric repeat sequences at this junction and the presence of microhomology at the junction site between these sequences are consistent with a role for nonhomologous end joining in formation of this junction.
Discussion
Our analysis of retrosequence formation in telomerase-negative yeast demonstrates that processed pseudogenes are commonly found at sites of chromosome rearrangements. Multiple studies have identified associations between retrotransposons and chromosome rearrangements, including studies in mammalian cells revealing that retrotransposons are often found at breakpoint junctions of translocations and deletions present in tumor cells or cells derived from individuals with inherited disorders (Abeysinghe et al. 2003; Florl and Schulz 2003). In addition, Ty1 cDNA sequences can be captured at sites of chromosome breaks repaired by homology-independent end-joining mechanisms (Teng et al. 1996; Yu and Gabriel 1999, 2004). However, our work raises the possibility that chromosome translocations and other rearrangements can be produced through the use of cDNA generated by the activity of endogenous retrotransposons for homology-dependent repair of chromosome breaks. These results emphasize the need to focus more attention on genetic and environmental conditions that promote formation of reverse transcripts and the potential use of reverse transcripts in the generation of chromosome rearrangements.
We showed that Ty1-mediated retrosequence formation is elevated and correlated with frequent chromosome rearrangements in telomerase-negative yeast. Analysis of four strains harboring processed pseudogenes and chromosome rearrangements indicates that processed pseudogenes are present at rearrangement junctions. Based on the role of RAD52 in retrosequence formation and the structures of sequences flanking processed pseudogenes, we propose that Ty1–ADH1HIS3–Ty1 cDNA molecules act as bridges for homologous recombination events between Ty1 sequences in different genomic locations. Transformation of yeast with exogenous DNA molecules designed to act as bridges for such recombination events has been shown to result in chromosome rearrangements (Tosato et al. 2005).
This study provides no direct evidence that retrosequence formation plays a role in telomere maintenance in the absence of telomerase. However, one breakpoint for each of three translocations we characterized was near or at a telomere. Therefore, the corresponding retrosequence insertions may have been associated with repair of chromosome breaks resulting from destabilized telomeres. It remains unclear whether the involvement of retrosequence formation in such repair events could somehow be advantageous to cells.
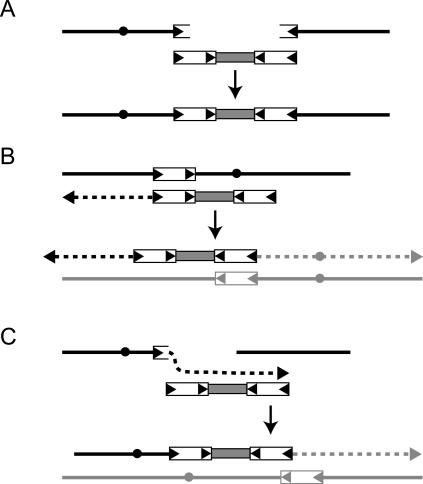
Based on our findings, we suggest that RAD52 has a role in processed pseudogene events and formation of chromosome rearrangements. We propose that retrosequences may be commonly incorporated through break-induced replication (BIR) or single-stranded annealing (SSA) events, since both of these processes are dependent on RAD52 and can occur in the other DNA repair mutants that we tested, including rad51Δ mutants (Malkova et al. 1996; Signon et al. 2001; Davis and Symington 2004; Krogh and Symington 2004). SSA events could involve Ty1 retrosequence cDNA molecules annealing to Ty1 sequences exposed by resection at sites of DNA damage to mediate repair (Fig. 5A). Insertion of Ty1–ADH1HIS3–Ty1 cDNA through SSA may have formed chromosome A (Fig. 4A). BIR involving Ty1 retrosequence cDNA could occur through variations of a two-step process (Mieczkowski et al. 2006). In one scenario, the cDNA itself could be extended by replication initiated at a chromosomal Ty1 sequence, followed by association of the Ty1 end of the resulting chromosome fragment with another chromosomal Ty1 sequence to initiate replication in the other direction (Fig. 5B). In a second scenario, the cDNA could be a template that is copied at a site of DNA damage, followed by invasion of another genomic site containing Ty1 sequences to complete the repair event (Fig. 5C). Chromosomes B, C, and D (Fig. 4B–D) were formed by nonreciprocal translocations that involved duplication of chromosome fragments, so these chromosomes may have been produced by BIR.
Figure 5.
Models for insertion of retrosequences by SSA or BIR. Black and gray lines represent chromosomes, dotted lines represent newly replicated DNA, white boxes with arrowheads represent Ty1, and gray boxes represent retrosequences. We show only one copy of Ty1 flanking either side of the retrosequence to simplify the drawing, even though our data were consistent with arrays of Ty1 flanking retrosequences at breakpoint junctions. (A) The drawing shows a broken chromosome with breakpoints in Ty1 sequences that is repaired by annealing of Ty1 retrosequence cDNA to each breakpoint. Drawings in B and C show alternative two-step BIR mechanisms for retrosequence insertion that result in nonreciprocal translocations. (B) Each end of Ty1 retrosequence cDNA is extended by separate BIR events that initiate from sites on two different chromosomes (Mieczkowski et al. 2006). (C) A breakpoint on one chromosome is extended by replication of Ty1 retrosequence cDNA, followed by invasion of a second site and replication of the corresponding chromosome fragment.
Our mapping of rearrangements was consistent with the presence of many copies of Ty1 flanking either side of retrosequences (Supplementary Fig. S3), but the mechanism responsible for formation of these arrays remains to be determined. Arrays of Ty1 elements have also been found at breakpoint junctions in other studies (Umezu et al. 2002). A detailed structural analysis of insertions of multimeric Ty1 arrays has been previously reported, but the structures identified could not conclusively indicate the mechanism responsible for array formation (Weinstock et al. 1990). Weinstock et al. (1990) proposed that array formation could involve aberrant-priming events during reverse transcription, recombination events between unintegrated cDNA intermediates, or recombination events between newly integrated Ty1 elements and unintegrated cDNA intermediates.
Our results concerning the role of RAD52 in retrosequence formation are similar to results obtained using a chromosomal ura2 donor allele (Schacherer et al. 2005), but they differ from the results of previous studies using pGALhis3AI and pGAL1his3ΔATG plasmid donor constructs (Derr and Strathern 1993; Derr 1998). These latter studies demonstrated that RAD52 and RAD1 are required for cDNA-mediated gene conversion of a chromosomal allele, but loss of RAD52 did not reduce the overall rate of retrosequence formation, whereas loss of RAD1 did reduce the rate. A role for Ty1 integrase in retrosequence formation was suggested to explain the lack of influence of RAD52 on the overall rate of retrosequence formation (Derr and Strathern 1993). We did not observe any ADH1his3AI conversion events in rad52Δ mutants, but loss of RAD52 did significantly reduce the frequency of ADH1HIS3 retrosequence formation, whereas loss of RAD1 did not have any influence on the overall frequency or the relative fraction of ADH1his3AI conversion events. Differences in strains or in retrosequence donors used might account for some differences in our results. It is also possible that use of the pGALhis3AI donor results in preferential formation of retrosequence cDNA with structures that are more amenable to insertion by Ty1 integrase, resulting in RAD52-independent processed pseudogene events. Ty1 integrase could play a role in retrosequence formation in our donor system, but it either plays a minor role, or its role is RAD52 dependent.
We observed that the overall frequency of retrosequence formation and the relative fraction of processed pseudogene events were elevated in telomerase-negative cells. Since processed pseudogenes are flanked by Ty1 sequences, this increase in the relative fraction of processed pseudogene events could result from elevated cDNA-mediated recombination with genomic Ty1 sequences in telomerase-negative yeast. The presence of flanking Ty1 regulatory sequences could result in expression of processed pseudogenes. We also found that a significant number of ADH1HIS3 retrosequences were fused to sequences of other ORFs, and such fusions could produce chimeric retrogenes. Ty1-mediated retrosequence formation could therefore play an important role in the generation of genes with novel functions and expression patterns, especially when cells are experiencing some form of genome maintenance stress. No gene relics in S. cerevisiae have been reported to have the structure of processed pseudogenes (Lafontaine et al. 2004), but this could be due to rapid degeneration of the Ty1 sequences flanking processed pseudogenes to the point that the Ty1 sequences are unrecognizable.
Based on these findings, we propose that reverse transcripts and retrotransposons play active roles in the formation of genome rearrangements. Moreover, these observations may have important implications for understanding mechanisms that produce chromosome rearrangements in other organisms, including humans, and for understanding the consequences of retrotransposon regulation by host factors.
Materials and methods
Yeast strains and media
Yeast strains were grown using standard media. Strains were derivatives of BY4742 (S288c background) or JC2980 (MATα ade2∷hisG his3Δ200 leu2∷hisG trp1∷hisG ura3-167), a strain from the GRF167 background. Details of strain constructions are provided with the Supplemental Material.
Generation and growth of telomerase-negative strains
We generated telomerase-negative derivatives of strains in the S288c background harboring the tlc1∷LEU2 allele and plasmid pRS317TLC1 (Teng and Zakian 1999), containing the wild-type TLC1 allele, by segregating the plasmid. Telomerase-negative derivatives of strains in the GRF167 background harboring an EST2–URA3–est2Δ1 construct at the EST2 locus were generated by selecting for Ura− cells using 5-fluoroorotic acid and by PCR screening for those in which est2Δ1, but not EST2, was retained. EST2 derivatives were used as telomerase-positive controls. Serial subculture of telomerase-negative strains through senescence and survivor formation was accomplished by streaking on YPD medium. Cells were grown for 3 d at 30°C for each subculture, and each subculture corresponded to ∼20–25 generations.
Assays for retrosequence formation
We measured the frequency of retrosequence formation as the frequency of His+ prototroph formation in strains containing ADH1his3AI. Cells from clusters of single colonies or patches of single colonies were inoculated into three cultures each at a density of 0.5 × 105 or 1 × 105 cells per milliliter in YPD broth and grown for 3 d at 20°C. The cell density of each culture was determined by spreading aliquots of 0.001–0.2 μL onto YPD agar, followed by incubation for 4 d at 30°C. Appropriate volumes of each culture were divided among SC-His plates, which were incubated for 7 d at 30°C. The number of His+ prototrophs formed in each set of triplicate cultures was divided by the total number of viable cells spread on SC-His medium to obtain a single His+ frequency value. The effect of galactose-induced expression of pGAL1Ty1kanAI in strains with plasmid pBJC546 was tested following the same procedure, except that cells were grown in SC-Ura broth with 2% glucose or SC-Ura broth with 1.5% galactose and 0.5% raffinose. After 3 d of growth, a one-third volume of YP broth was added and the cultures were incubated for one additional day at 20°C. Cells were spread onto SC-Ura and SC-Ura-His plates, which were incubated for 7 d at 30°C, to determine the His+ frequency in cells that retained the plasmid throughout the experiment.
Preparation of genomic DNA and agarose-embedded chromosomal DNA
Genomic DNA was prepared using a zymolyase-based procedure, as described previously (Adams et al. 1997), with the addition of a phenol/chloroform extraction step prior to the final isopropanol precipitation. Intact yeast chromosomal DNA embedded in agarose was prepared from 1 mL or 2–4 mL of overnight cultures grown in YPD or SC-His medium, respectively (Gerring et al. 1991). For restriction digestions of agarose-embedded DNA, agarose plugs were washed in three changes of 10 mM Tris (pH 7.5). After equilibrating the plugs in three changes of the appropriate restriction enzyme buffer, samples were incubated with 240 U/mL restriction enzyme in a volume of 300 μL (agarose plug volumes were ∼100 μL) for 16–20 h at 37°C. Digested samples were immediately resolved by pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis and Southern blotting
Intact yeast chromosomes or chromosome restriction fragments were resolved on 1% agarose gels (SeaKem Gold, Lonza) in 0.5× TBE buffer by pulsed-field gel electrophoresis using a CHEF gel electrophoresis system (Bio-Rad CHEF-DR II). Gels were run for 24 h at 6 V/cm with pulse times of 45–115 sec or for 21 h with pulse times of 1–25 sec for intact chromosomes or restriction fragments, respectively. Southern blots of chromosomes or chromosome restriction fragments resolved on CHEF gels were performed as described previously (Maxwell et al. 2004). Riboprobe templates for ORF sequences used for mapping chromosome rearrangement junctions were generated by PCR with primers that incorporated T7 promoter sequences. These primer sequences are available on request.
Competitive PCR to quantify retrosequence cDNA
Retrosequence cDNA was quantified relative to a single-copy gene using a previously described strategy (Maxwell et al. 2004), and the details of this method are given in the Supplemental Material.
PCR, inverse PCR, cloning, and sequencing
These methods are described in the Supplemental Material, and primer sequences are listed in Supplementary Table S2.
Statistical analyses
Median retrosequence formation frequency values obtained from different data sets were compared for significant differences using the Mann-Whitney rank sum test. The mean numbers of chromosome rearrangements for control isolates lacking retrosequences and test isolates harboring ADH1HIS3 retrosequences were compared using the Student’s t-test. Distributions of His+ events between different classes in different strains (Table 2) were compared using the χ2 test.
Acknowledgments
We thank C. Coros and T. Petes for comments on the manuscript and the Wadsworth Center Molecular Genetics Core for DNA sequencing. This work was supported by National Institutes of Health grant GM52072 to M.J.C.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1604707
References
- Abeysinghe S.S., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N., Krawczak M., Ball E.V., Cooper D.N., Ball E.V., Cooper D.N., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Adams A., Gottschling D.E., Kaiser C.A., Stearns T., Gottschling D.E., Kaiser C.A., Stearns T., Kaiser C.A., Stearns T., Stearns T. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- Boeke J.D., Styles C.A., Fink G.R., Styles C.A., Fink G.R., Fink G.R. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol. Cell. Biol. 1986;6:3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M.J., Garfinkel D.J., Garfinkel D.J. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Symington L.S., Symington L.S. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr L.K. The involvement of cellular recombination and repair genes in RNA-mediated recombination in Saccharomyces cerevisiae. Genetics. 1998;148:937–945. doi: 10.1093/genetics/148.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr L.K., Strathern J.N., Strathern J.N. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- Derr L.K., Strathern J.N., Garfinkel D.J., Strathern J.N., Garfinkel D.J., Garfinkel D.J. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- Dunham M.J., Badrane H., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D., Badrane H., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D., Adams J., Brown P.O., Rosenzweig F., Botstein D., Brown P.O., Rosenzweig F., Botstein D., Rosenzweig F., Botstein D., Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T., Maestre J., Heidmann T., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Florl A.R., Schulz W.A., Schulz W.A. Peculiar structure and location of 9p21 homozygous deletion breakpoints in human cancer cells. Genes Chromosomes Cancer. 2003;37:141–148. doi: 10.1002/gcc.10192. [DOI] [PubMed] [Google Scholar]
- Gerring S.L., Connelly C., Hieter P., Connelly C., Hieter P., Hieter P. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 1991;194:57–77. doi: 10.1016/0076-6879(91)94007-y. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Feldser D.M., Greider C.W., Feldser D.M., Greider C.W., Greider C.W. Telomere dysfuntion increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- Harrison P.M., Echols N., Gerstein M.B., Echols N., Gerstein M.B., Gerstein M.B. Digging for dead genes: An analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res. 2001;29:818–830. doi: 10.1093/nar/29.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.K., Bennetzen J.L., Bennetzen J.L. Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell. 1994;6:1177–1186. doi: 10.1105/tpc.6.8.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lafontaine I., Fischer G., Talla E., Dujon B., Fischer G., Talla E., Dujon B., Talla E., Dujon B., Dujon B. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene. 2004;335:1–17. doi: 10.1016/j.gene.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Lemoine F.J., Degtyareva N.P., Lobachev K., Petes T.D., Degtyareva N.P., Lobachev K., Petes T.D., Lobachev K., Petes T.D., Petes T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase: A model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E.H., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Malkova A., Ivanov E.L., Haber J.E., Ivanov E.L., Haber J.E., Haber J.E. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.H., Coombes C., Kenny A.E., Lawler J.F., Boeke J.D., Curcio M.J., Coombes C., Kenny A.E., Lawler J.F., Boeke J.D., Curcio M.J., Kenny A.E., Lawler J.F., Boeke J.D., Curcio M.J., Lawler J.F., Boeke J.D., Curcio M.J., Boeke J.D., Curcio M.J., Curcio M.J. Ty1 mobilizes subtelomeric Y′ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol. Cell. Biol. 2004;24:9887–9898. doi: 10.1128/MCB.24.22.9887-9898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P.A., Lemoine F.J., Petes T.D., Lemoine F.J., Petes T.D., Petes T.D. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Ohshima K., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N., Yada T., Gojobori T., Sakaki Y., Okada N., Gojobori T., Sakaki Y., Okada N., Sakaki Y., Okada N., Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C.D., Pennaneach V., Kolodner R.D., Pennaneach V., Kolodner R.D., Kolodner R.D. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 2005;25:7226–7238. doi: 10.1128/MCB.25.16.7226-7238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A.J., Shafer B.K., Garfinkel D.J., Shafer B.K., Garfinkel D.J., Garfinkel D.J. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics. 2000;154:543–556. doi: 10.1093/genetics/154.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Tourrette Y., Souciet J.L., Potier S., De Montigny J., Tourrette Y., Souciet J.L., Potier S., De Montigny J., Souciet J.L., Potier S., De Montigny J., Potier S., De Montigny J., De Montigny J. Recovery of a function involving gene duplication by retroposition in Saccharomyces cerevisiae. Genome Res. 2004;14:1291–1297. doi: 10.1101/gr.2363004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., de Montigny J., Welcker A., Souciet J.L., Potier S., de Montigny J., Welcker A., Souciet J.L., Potier S., Welcker A., Souciet J.L., Potier S., Souciet J.L., Potier S., Potier S. Duplication processes in Saccharomyces cerevisiae haploid strains. Nucleic Acids Res. 2005;33:6319–6326. doi: 10.1093/nar/gki941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D.T., Banerjee M., Bowen B., Curcio M.J., Banerjee M., Bowen B., Curcio M.J., Bowen B., Curcio M.J., Curcio M.J. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D.T., Kenny A.E., Gamache E.R., Mou Z., Curcio M.J., Kenny A.E., Gamache E.R., Mou Z., Curcio M.J., Gamache E.R., Mou Z., Curcio M.J., Mou Z., Curcio M.J., Curcio M.J. Activation of a LTR-retrotransposon by telomere erosion. Proc. Natl. Acad. Sci. 2003;100:15736–15741. doi: 10.1073/pnas.2136609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signon L., Malkova A., Naylor M.L., Klein H., Haber J.E., Malkova A., Naylor M.L., Klein H., Haber J.E., Naylor M.L., Klein H., Haber J.E., Klein H., Haber J.E., Haber J.E. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.C., Zakian V.A., Zakian V.A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.C., Kim B., Gabriel A., Kim B., Gabriel A., Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- Torrents D., Suyama M., Zdobnov E., Bork P., Suyama M., Zdobnov E., Bork P., Zdobnov E., Bork P., Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosato V., Waghmare S.K., Bruschi C.V., Waghmare S.K., Bruschi C.V., Bruschi C.V. Non-reciprocal chromosomal bridge-induced translocation (BIT) by targeted DNA integration in yeast. Chromosoma. 2005;114:15–27. doi: 10.1007/s00412-005-0332-x. [DOI] [PubMed] [Google Scholar]
- Umezu K., Hiraoka M., Mori M., Maki H., Hiraoka M., Mori M., Maki H., Mori M., Maki H., Maki H. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: Retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics. 2002;160:97–110. doi: 10.1093/genetics/160.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang L., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E., Hieter P., Vogelstein B., Kinzler K.W., Zhang L., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E., Hieter P., Vogelstein B., Kinzler K.W., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E., Hieter P., Vogelstein B., Kinzler K.W., Vogelstein J., Basrai M.A., Bassett D.E., Hieter P., Vogelstein B., Kinzler K.W., Basrai M.A., Bassett D.E., Hieter P., Vogelstein B., Kinzler K.W., Bassett D.E., Hieter P., Vogelstein B., Kinzler K.W., Hieter P., Vogelstein B., Kinzler K.W., Vogelstein B., Kinzler K.W., Kinzler K.W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Wang W., Zheng H., Fan C., Li J., Shi J., Cai Z., Zhang G., Liu D., Zhang J., Vang S., Zheng H., Fan C., Li J., Shi J., Cai Z., Zhang G., Liu D., Zhang J., Vang S., Fan C., Li J., Shi J., Cai Z., Zhang G., Liu D., Zhang J., Vang S., Li J., Shi J., Cai Z., Zhang G., Liu D., Zhang J., Vang S., Shi J., Cai Z., Zhang G., Liu D., Zhang J., Vang S., Cai Z., Zhang G., Liu D., Zhang J., Vang S., Zhang G., Liu D., Zhang J., Vang S., Liu D., Zhang J., Vang S., Zhang J., Vang S., Vang S., et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock K.G., Matrangelo M.F., Burkett T.J., Garfinkel D.J., Strathern J.N., Matrangelo M.F., Burkett T.J., Garfinkel D.J., Strathern J.N., Burkett T.J., Garfinkel D.J., Strathern J.N., Garfinkel D.J., Strathern J.N., Strathern J.N. Multimeric arrays of the yeast retrotransposon Ty. Mol. Cell. Biol. 1990;10:2882–2892. doi: 10.1128/mcb.10.6.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Gabriel A., Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- Yu X., Gabriel A., Gabriel A. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166:741–751. doi: 10.1534/genetics.166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M., Harrison P.M., Liu Y., Gerstein M., Liu Y., Gerstein M., Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Carriero N., Gerstein M., Carriero N., Gerstein M., Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]