Abstract
In the budding yeast Saccharomyces cerevisiae, microtubule-organizing centers called spindle pole bodies (SPBs) are embedded in the nuclear envelope, which remains intact throughout the cell cycle (closed mitosis). Kinetochores are tethered to SPBs by microtubules during most of the cell cycle, including G1 and M phases; however, it has been a topic of debate whether microtubule interaction is constantly maintained or transiently disrupted during chromosome duplication. Here, we show that centromeres are detached from microtubules for 1–2 min and displaced away from a spindle pole in early S phase. These detachment and displacement events are caused by centromere DNA replication, which results in disassembly of kinetochores. Soon afterward, kinetochores are reassembled, leading to their recapture by microtubules. We also show how kinetochores are subsequently transported poleward by microtubules. Our study gives new insights into kinetochore–microtubule interaction and kinetochore duplication during S phase in a closed mitosis.
[Keywords: Kinetochore, microtubule, S phase, Saccharomyces cerevisiae, closed mitosis]
To maintain genetic integrity, eukaryotic cells must segregate their chromosomes properly to opposite spindle poles prior to cell division. Sister chromatid segregation mainly depends on the forces generated by microtubules, which extend from microtubule-organizing centers (MTOCs) and attach to kinetochores, large protein complexes assembled at centromeres on chromosomes (Maiato et al. 2004; Kline-Smith et al. 2005; T.U. Tanaka et al. 2005). In most metazoan cells, MTOCs (called centrosomes) are located outside the nuclear envelope during interphase and, only after the nuclear envelope breaks down in early M phase (prometaphase), can microtubules from MTOCs access and capture kinetochoes (open mitosis) (Bornens 2002; Sazer 2005). In contrast, in many unicellular eukaryotes such as yeasts, MTOCs (called spindle pole bodies; SPBs) are embedded in the nuclear envelope, which remains intact throughout the cell cycle (closed mitosis) (Jaspersen and Winey 2004; Sazer 2005). Thus, microtubules from MTOCs may interact with kinetochores before M phase in a closed mitosis. However, it is still elusive how kinetochores interact with microtubules during each cell cycle phase in a closed mitosis, and which aspects of kinetochore–microtubule interaction are similar and dissimilar between an open and closed mitosis.
Budding yeast Saccharomyces cerevisiae is a well-studied model organism that undergoes a closed mitosis (Winey and O’Toole 2001; T.U. Tanaka et al. 2005). In this organism, centromeres are tethered to spindle poles by microtubules during most of the cell cycle, including G1 and M phases (Guacci et al. 1997; Jin et al. 2000; Winey and O’Toole 2001; Tanaka et al. 2002) (we propose that there is no G2 phase in this organism; see Discussion). Indeed, it has been thought that centromeres might never detach from microtubules throughout the cell cycle (see Discussion in Winey and O’Toole 2001). If this were the case, how would kinetochores be duplicated upon centomere DNA replication in S phase? In order to maintain kinetochore–microtubule attachment, kinetochores may have to be duplicated in a conservative manner similarly to SPBs and centrioles in metazoan cells (Adams and Kilmartin 2000; Pereira et al. 2001; Stearns 2001); i.e., the old one, inherited from the previous cell cycle, remains intact, while the new one is generated in the present cycle.
However, recent indirect evidence has suggested that kinetochores might be transiently disassembled during S phase, causing centromeres to detach from microtubules (Pearson et al. 2004; Tanaka 2005; K. Tanaka et al. 2005); nonetheless, this process has not yet been directly proven. If centromeres indeed detach from microtubules in S phase, direct visualization of this process would help its characterization.
Here, we demonstrate that centromeres remain adjacent to a spindle pole in G1, but detach from microtubules and move away from a spindle pole for 1–2 min when the centromere DNA replicates during S phase. Subsequently, centromeres are recaptured by microtubules and transported to the vicinity of a spindle pole. Such centromere detachment from microtubules is dependent on DNA replication and caused by transient kinetochore disassembly at centomeres. These processes have not been visualized previously because of their extremely transient nature. Our data resolve a long-standing controversy on the status of kinetochore–microtubule interaction during S phase in S. cerevisiae, and give crucial insights into early kinetochore–microtubule interaction that eventually ensures high-fidelity chromosome segregation in the subsequent anaphase.
Results
Centromeres are transiently detached from microtubules and move away from a spindle pole during S phase
To study centromere behavior with time-lapse microscopy, we marked CEN5 and CEN15 by the adjacent insertion of a tet or lac operator array, respectively. These arrays were bound by Tet repressors fused with three tandem copies of cyan fluorescent proteins (CFP) and by LacI with a single copy of green fluorescent protein (GFP); thus, CEN5 and CEN15 were visualized as small CFP and GFP dots, respectively. Microtubules were also visualized by expression of α-tubulin (TUB1) fused with yellow fluorescent protein (YFP). Using the JP3 filter set (see Materials and Methods), CFP/GFP and YFP were separately visualized, and the two CENs could be distinguished because CEN15-GFP showed higher intensity than CEN5-CFP.
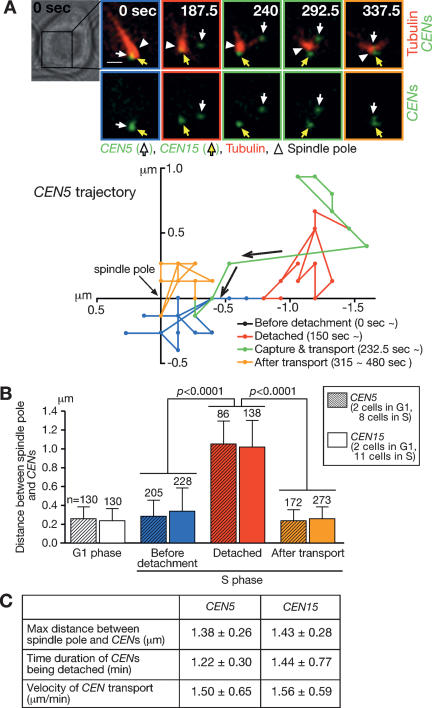
We synchronized the cell cycle of cells with α-factor treatment and subsequently released them into fresh medium. Both CEN5 and CEN15 stayed in the vicinity of a spindle pole (<0.5 μm from the center of the pole) both during G1 phase (Fig. 1B, white bars) and presumed entry into S phase (Fig. 1A,B, blue). However, just before bud emergence, which corresponds to early S phase (see below), both CENs detached from microtubules and moved away from a spindle pole (Fig. 1A,B [red in A shows CEN5 detachment]); note that SPBs have not yet separated and cells have a single spindle pole in S phase (Lim et al. 1996). It was very unlikely that CEN “detachment” from microtubules was an artifactual observation because (1) we could visualize single microtubules by our imaging method (Supplementary Note 1), and (2) CENs moved away from a pole coincidentally upon detachment. While CENs were detached, their distance from a spindle pole was 1.0 μm on average and was as much as 1.4 μm (Fig. 1B [red], C). After CENs were detached for 1.2–1.4 min on average (Fig. 1C), they were recaptured by microtubules and subsequently transported to the vicinity of a spindle pole at an average velocity of 1.5 μm/min (Fig. 1A [green], C), where they stayed thereafter (Fig. 1A,B, orange).
Figure 1.
Centromeres are transiently detached from microtubules and are displaced away from a spindle pole during S phase. CEN5-tetOs TetR-3CFP CEN15-lacOs GFP-LacI YFP-TUB1 cells (T4243) were treated with α-factor and subsequently released to fresh medium. After 30 min, CFP/GFP and YFP images were collected every 7.5 sec for 8 min. (A, top) Representative time-lapse images show CEN5 and CEN15 in green and microtubules in red. White arrows, yellow arrows, and white arrowheads indicate CEN5, CEN15, and a spindle pole, respectively. Time is shown in seconds in the montage (0 sec: start of image acquisition). Bar, 1 μm. (Bottom) The trajectory of CEN5 (its position relative to a spindle pole) in the same cell shown at the top is plotted along X- and Y-axes before detachment from microtubules (blue), while being detached (red), after recapture by microtubules/during transport toward a spindle pole (green) and after transport (orange; the same colors were also used to outline frames of the montage on the top). Arrows indicate the direction of CEN5 motion. (B) The distance between CEN5/CEN15 and a spindle pole was measured at each time point in two cells in G1 phase (5–13 min after release from α-factor arrest) and eight and 11 cells in S phase, where CEN5 and CEN15 were detached from microtubules, respectively. “n” denotes the number of time points of measurement. Error bars show SD. P values were obtained by comparing indicated values, separately for CEN5 and CEN15, using an unpaired t-test. (C) For CEN5 and CEN15, their maximum distance from a spindle pole (while CEN was detached from microtubules), duration of CEN detachment, and the velocity of CEN transport (mean ± SD) are shown; the data set obtained in B was analyzed.
One might predict that CENs could move around freely by diffusion in the nucleus while being detached from microtubules. However, we found that they showed more restricted motion than expected from free diffusion (Supplementary Fig. S1). Such restriction was previously reported for S-phase motion of replication origins (Heun et al. 2001) and was perhaps due to replication forks (around CENs) being associated with replication factories during S phase (Kitamura et al. 2006).
Centromere detachment from microtubules coincides with its DNA replication
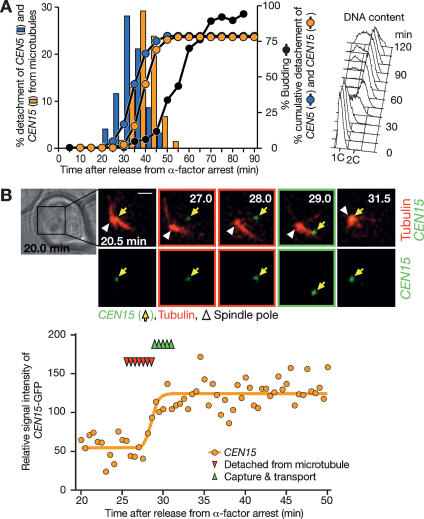
Next, we determined timing of transient detachment of CEN5 and CEN15 in the cell cycle after release from α-factor arrest. To reduce photobleaching of YFP-Tub1, we changed the field of observation every 5 min, and the percentage of CEN detachment was scored during each 5-min interval (Fig. 2) (cells that had already shown detachment from the beginning of each interval were not counted). Frequency of CEN5 and CEN15 detachment showed a peak at 30–35 and 35–40 min, after release from α-factor arrest, respectively (Fig. 2A, left). This was shortly after the time that we could detect DNA replication by FACS analysis (Fig. 2A, right).
Figure 2.
Centromere detachment from microtubules coincides with its DNA replication. (A) Centromere detachment from microtubules occurs in early S phase. T4243 cells (see legend for Fig. 1) were treated with α-factor and subsequently released to fresh medium. CFP/GFP and YFP images were collected every 7.5 sec using the JP3 filter set (see Materials and Methods). To reduce photobleaching of YFP-Tub1 signals, the field of microscopic observation was changed every 5 min, and the percentage of CEN5 (blue) and CEN15 (orange) detachment was scored during each 5-min interval (bars). The cumulative percentage of CEN detachment is shown as lines. The percentage of cells with buds (line with black dots) and FACS DNA content (right) are also shown. (B) Centromere detachment and its DNA replication are coincidental. CEN15-lacOs GFP-LacI YFP-TUB1 cells (T5276) were treated with α-factor and subsequently released to fresh medium. After 20 min, GFP and YFP images were collected every 30 sec for 30 min. (Top) Representative time- lapse images show CEN15-GFP in green and microtubules in red. Yellow arrows and white arrowheads indicate CEN15 and a spindle pole, respectively. Time indicated on images: minutes after release from α-factor arrest. Bar, 1 μm. (Bottom) The intensity of a CEN15-GFP dot was measured and plotted, and its change was approximated by a regression curve (orange) with the method described previously (Kitamura et al. 2006). Red and green triangles show the time points at which CEN15 was detached from microtubules and subsequently reassociated with them (until its return to the vicinity of a spindle pole by transport), respectively (the same colors were also used to outline frames of the montage on the top).
Because centomere DNAs replicate in early S phase (McCarroll and Fangman 1988), we predicted that centromeres might detach from microtubules upon their DNA replication. This prediction was consistent with the result from a genome-wide replication timing study that found that a CEN5 DNA replicates 4 min earlier than CEN15 DNA (Raghuraman et al. 2001; Supplementary Note 2). To test this prediction directly, we evaluated the timing of CEN15 DNA replication with live-cell imaging in the same cells in which we also observed CEN15 detachment from microtubules (Fig. 2B; Supplementary Fig. S2). This evaluation was based on our recent finding that DNA replication timing of a chromosome locus coincided with the increase in the intensity of the GFP-LacI dot on a lac operator array inserted at this locus (Kitamura et al. 2006). We found that CEN15 detached from microtubules when its dot intensity increased (at mid-point of the increase on the regression curve), or up to 3 min earlier (Supplementary Note 3), suggesting that centromere detachment indeed happened upon its DNA replication.
Centromere detachment from microtubules is dependent on its DNA replication
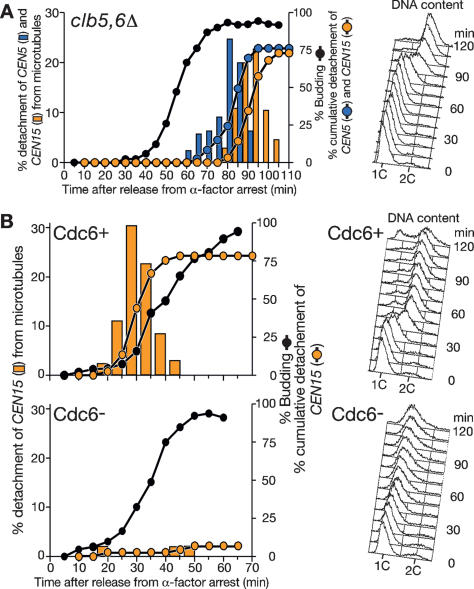
We then addressed whether CEN detachment from microtubules is dependent on CEN DNA replication. When B-type cyclins CLB5 and CLB6 were deleted, DNA replication was significantly delayed relative to bud emergence (Schwob and Nasmyth 1993), and in such cells, the timing of CEN5 and CEN15 detachment was also delayed relative to bud emergence, but still happened when DNA replication had started (Fig. 3A; cf. Fig. 2A).
Figure 3.
Centromere detachment from microtubules is dependent on DNA replication. (A) The timing of centromere detachment is delayed relative to bud emergence when DNA replication is delayed due to deletion of CLB5,CLB6. clb5Δ clb6Δ CEN5-tetOs TetR-3CFP CEN15-lacOs GFP-LacI YFP-TUB1 cells (T5115) were treated and images were acquired and analyzed as in Figure 2A. Symbols and colors are as in Figure 2A. (B) Cdc6-depleted cells rarely show centromere detachment from microtubules. Pgal-CDC6 cdc6Δ CEN15-lacOs GFP-LacI YFP-TUB1 cells (T5118) were grown in medium containing galactose and raffinose, arrested with nocodazole treatment, released to medium containing glucose and α-factor, then subsequently rereleased to fresh medium containing glucose, as described previously (Severin et al. 2001). Images were acquired and analyzed as in Figure 2A. Symbols and colors are as in Figure 2A.
We next studied whether CEN detached from microtubules in Cdc6-depleted cells. Cdc6 associates with DNA replication origins (Tanaka et al. 1997) and is required for DNA replication initiation (Blow and Tanaka 2005). Cdc6-depleted cells do not replicate DNA, but still undergo other cell cycle events such as bud emergence and bipolar spindle formation (Piatti et al. 1995). We inhibited CDC6 expression in cells where the only CDC6 was under control of a galactose-inducible promoter (Piatti et al. 1996). When CDC6 expression was suppressed and DNA replication inhibited, CEN15 rarely detached from microtubules either before or during bud emergence (Fig. 3B). Taken together, we concluded that CEN detachment from microtubules was dependent on its DNA replication.
The majority of centromeres show evidence of detachment only once during S phase
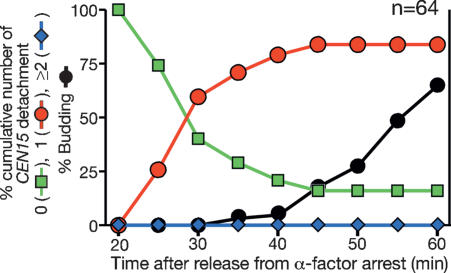
If centromeres detach from microtubules only upon their DNA replication, detachment should happen only once for each CEN in an individual cell during the whole S phase. To test this, we investigated possible CEN15 detachment focusing on a group of cells for a long period (40 min), starting 20 min after release from α-factor arrest (Fig. 4); this period should have covered most, if not all, detachment events (see Fig. 2A). The majority of cells showed CEN15 detachment from microtubules just prior to bud emergence and most of them showed detachment only once during observation (Fig. 4). The cumulative percentage of CEN that had undergone detachment reached 84%; a similar cumulative percent (CEN5, 79%; CEN15, 78%) was also obtained in Figure 2A. Thus, the majority of centromeres showed evidence of detachment only once in each cell during the whole S phase. Given that we set rather rigorous criteria for counting CEN detachment to avoid false judgement (Materials and Methods; Supplementary Note 4), we assume that, in virtually all cells, CEN detachment from microtubules happened during S phase.
Figure 4.
The majority of centromeres show detachment only once during S phase. CEN15-lacOs GFP-LacI GFP-TUB1 cells (T5280) were treated with α-factor and subsequently released to fresh medium. After 20 min, GFP images were collected every 20 sec for 40 min. The colored lines show the percentage of cells with the following cumulative number of CEN15 detachment from microtubules: zero (green), once (red,) and twice or more (blue). The black line shows the percentage of cells with buds.
DNA replication and Aurora kinase Ipl1 facilitate centromere detachment from microtubule, independently of each other
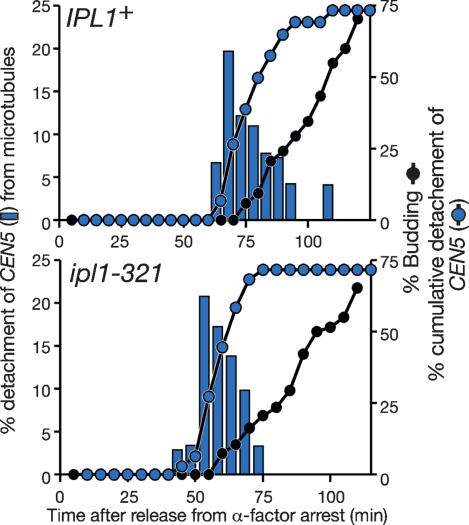
We previously suggested that Ipl1, the ortholog of Aurora B kinase in metazoan cells, promotes reorientation of kinetochore–microtubule interaction in budding yeast (Tanaka et al. 2002; Dewar et al. 2004). We proposed that this reorientation occurred in a tension-dependent manner, thus achieving proper sister kinetochore biorientation on a bipolar mitotic spindle. Indeed, the majority of ipl1-321 mutant cells showed defects in this reorientation and had mono-oriented sister kinetochores on a bipolar spindle at 35°C, the restrictive temperature for this mutant (Biggins et al. 1999; Tanaka et al. 2002). To resolve syntelic attachment (i.e., attachment of both sister kinetochores to microtubules from the same pole) (T.U. Tanaka et al. 2005), Aurora B/Ipl1 must facilitate detachment of at least one sister kinetochore from microtubules to achieve proper biorientation.
Given that DNA replication also causes centromere detachment from microtubules, an intriguing question is whether Ipl1 also regulates this replication-dependent detachment. To address this, we investigated CEN5 detachment in wild-type and ipl1-321 cells at 35°C. In both cells, CEN5 transiently detached from microtubules and moved away from a spindle pole, with similar frequency and timing (relative to budding), in the beginning of S phase (Fig. 5). The period of CEN5 detachment and the maximum distance of CEN5 from a spindle pole while detached were also similar between the two cells (data not shown). Thus, Ipl1 function is not required for centromere detachment from microtubules caused by DNA replication. Moreover, because Ipl1 can promote reorientation of unreplicated centromeres (Tanaka et al. 2002; Dewar et al. 2004), Ipl1 should be able to promote centromere detachment independently of DNA replication. Taken together, Ipl1 and DNA replication both facilitate centromere detachment from microtubules, but independently of each other.
Figure 5.
DNA replication causes centromere detachment from microtubules independently of Aurora kinase Ipl1. IPL1+ (T5052) and ipl1-321 (T5429) cells with CEN5-tetOs TetR-3CFP GFP-TUB1 were treated as in Figure 2A, except that temperature for cell culture was shifted to 35°C when cells were released from α-factor arrest. CFP and GFP images were acquired as in Figure 2A, but at 35°C and using the JP4 filter set (see Materials and Methods). Symbols and colors are as in Figure 2A.
Transient kinetochore disassembly upon centromere DNA replication
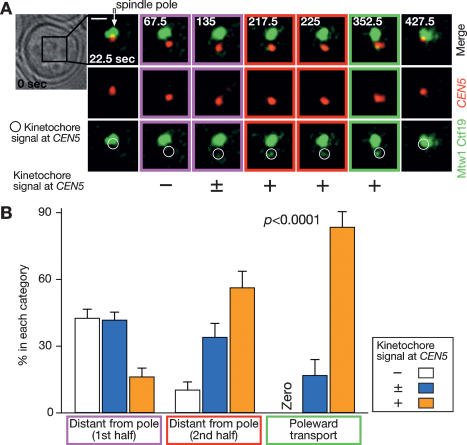
How does DNA replication cause centromere detachment from microtubules and its recapture within a short period? A simple explanation would be that centromere DNA replication causes kinetochore disassembly, leading to centromere detachment, and subsequent kinetochore reassembly allows its recapture by microtubules. To address this, we evaluated the signal intensity of kinetochore components Mtw1 and Ctf19 (Ortiz et al. 1999; Goshima and Yanagida 2000), each fused with four tandem copies of GFP, which colocalized at CFP-labeled CEN5 (Fig. 6; Supplementary Fig. S3). The GFP signal was not detected at CEN5 when it moved away from a spindle pole, but the GFP signal was subsequently detected before CEN5 returned to the vicinity of a spindle pole (Fig. 6A).
Figure 6.
Transient kinetochore disassembly upon centromere DNA replication. MTW1-4GFP CTF19-4GFP CEN5-tetOs TetR-3CFP cells (T5529) were treated with α-factor and subsequently released to fresh medium. After 25 min, CFP and GFP images were collected every 7.5 sec for 10 min. (A) Representative time-lapse images show CEN5-CFP in red and Mtw1-4GFP/Ctf19-4GFP in green. Time is shown in seconds in the montage (0 sec: start of image acquisition). White arrows and circles indicate the positions of a spindle pole and CEN5, respectively. The intensity of GFP signals at CEN5 was quantified and scored as “−” “±,” and “+,” as described in Materials and Methods. Colors outlining frames of the montage denote categories quantified in B (see below). Bar, 1 μm. (B) The GFP intensity was scored at each time point in 10 individual cells, and the mean (±SE) percentage of the 10 cells for each scoring category was depicted during the following three periods: “distant from pole, 1st half” (purple) and “distant from pole, 2nd half” (red), which equally divided the period of CEN5 being displaced from a spindle pole (CEN5–spindle pole distance >0.7 μm; during which we estimate CEN5 was detached from microtubules), and “poleward transport” (green), during which CEN5 moved toward a spindle pole. The data from each individual cell are shown in Supplementary Figure S3. The P value was obtained by testing the possible difference between the three periods using the GFP signal-intensity data together from the 10 individual cells (rather than the averaged percentages), using Kruskal-Wallis nonparametric test.
We then scored the intensity of GFP signals at CEN5. Soon after CEN5 moved away from a spindle pole (the first half of “distant from pole”; Fig. 6, framed in purple), discernible GFP signals (Fig. 6B, bars in orange) of the kinetochore components were found at CEN5 in only 16% of the time points. Afterward (the second half of “distant from pole”; Fig. 6, framed in red), GFP signals were observed more frequently (56%). When CEN5 was subsequently transported poleward by microtubules (Fig. 6, framed in green), kinetochore component signals were observed at CEN5 even more frequently (83%).
In the above experiment, it was not possible to compare the amount of kinetochore components at individual centromeres in G1 and S phase, because centromeres were clustered in the vicinity of a spindle pole in G1 phase. To visualize kinetochore components at a single CEN in G1, we regulated the CEN activity by an adjacently inserted galactose-inducible promoter (Supplementary Fig. S4; Hill and Bloom 1987). Kinetochore component signals (Mtw1-4GFP and Ctf19-4GFP) were detected soon after the CEN (marked with CFP) was conditionally activated (before CEN reached the vicinity of a spindle pole by poleward transport) in G1 phase, but later in S phase their amount was reduced (and subsequently recovered) when the same CEN detached from microtubules in the same cell (Supplementary Note 5).
Collectively, our data suggest that kinetochores, at least their outer components (i.e., those supposedly close to microtubule attachment sites rather than to centromere DNA) (McAinsh et al. 2003) such as Mtw1 and Ctf19, are disassembled upon centromere DNA replication, leading to centromere detachment from microtubules; soon afterward, kinetochores are reassembled, causing centromere recapture by microtubules. Inner kinetochore components might be also disassembled and reassembled upon centromere DNA replication, considering that the centromere-specific histone H3 variant Cse4 shows turnover at centromeres specifically during S phase (Pearson et al. 2004).
Both lateral sliding and end-on pulling operate for centromere transport toward a spindle pole in normal S phase
How are centromeres recaptured by microtubules and subsequently transported toward a spindle pole? We recently found that centromeres were initially captured by the lateral surface of microtubules (K. Tanaka et al. 2005). Subsequent poleward movement occurs in two distinct ways: lateral sliding, in which centromeres move along the side of a microtubule, and microtubule end-on pulling, in which the centromere is tethered to the end of a microtubule and is pulled poleward as the microtubule shrinks (Tanaka et al. 2007). Kar3, a kinesin-14 family member, is essential to drive poleward lateral sliding, whereas the Dam1 complex is crucial for end-on pulling. Indeed, to promote this process, the Dam1 complex continuously colocalizes at a centromere during microtubule end-on pulling, while such continuous colocalization is not found during lateral sliding (Tanaka et al. 2007). These results were obtained when we regulated the centromere activity and turned it on in metaphase-arrested cells (centromere reactivation system) (K. Tanaka et al. 2005). In addition, in normal S phase, we also found that Kar3 and the Dam1 complex redundantly facilitated poleward centromere transport after its recapture by microtubules (Tanaka et al. 2007).
We can distinguish lateral sliding and end-on pulling using the centromere reactivation system; however, this has been a difficult task in normal S phase, where short microtubules frequently overlap with each other. Thus, it has been unclear whether both lateral sliding and end-on pulling operate for centromere transport in normal S phase or whether one mechanism works only as a backup of the other.
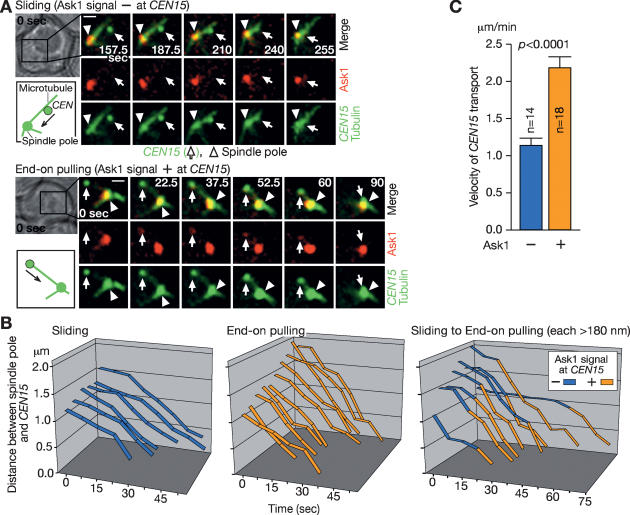
To address this, we distinguished the two processes by labeling the Dam1 complex component Ask1 with three tandem copies of CFP (Fig. 7A). We then scored continuous colocalization of CFP signals with GFP-labeled CEN15 when CEN15 was transported toward a spindle pole following displacement from a spindle pole in S phase. Ask1 colocalization was found during CEN15 transport (>180 nm toward a spindle pole) in 42% of cells (10 out of 24; judged as end-on pulling), while it was not detected in 25% of cells (six out of 24; judged as lateral sliding). In the remaining cells (33%; eight out of 24), Ask1-3CFP signals colocalized with CEN15, not in the beginning, but during the later phase of transport (judged as conversion from sliding to end-on pulling; Fig. 7B). We found no cells where colocalization was found in the beginning of transport, but then subsequently disappeared. Furthermore, CEN15 transport velocity was obviously higher during its colocalization with Ask1 (judged as end-on pulling) (Fig. 7C).
Figure 7.
Lateral sliding and end-on pulling for centromere transport in S phase. ASK1-3CFP CEN15-lacOs GFP-LacI GFP-TUB1 cells (T5363) were treated and CFP and GFP images were collected as in Figure 6. (A) Representative time-lapse images show Ask1-3CFP in red and CEN15-GFP/GFP-Tub1 in green. In the bottom image sequence, the Ask1 signal continuously colocalized, and in the top image sequence it did not. Time is shown in seconds in the montage (0 sec: start of image acquisition). White arrows and arrowheads indicate the positions CEN15 and a spindle pole, respectively. Bar, 1 μm. (B) Graphs show motion of CEN15 in each cell. CEN15–spindle pole distance was plotted as a function of time. “0 sec” indicates the start of CEN15 transport by end-on pulling or sliding. When the Ask1 signal continuously colocalized (orange) or not (blue) with CEN15 while being transported >180 nm, it was scored as CEN15 transport by end-on pulling or sliding, respectively. Note that if CEN15 did not colocalize with Ask1 and did not move >180 nm after it was reassociated with microtubules and before it was transported by end-on pulling (i.e., with Ask1 colocalization), such a phase was not scored as sliding and was not included in these graphs. (C) Graphs showing the velocity (mean ± SE) of CEN15 transport by end-on pulling (orange) or sliding (blue) toward a spindle pole.
If scoring Ask1 colocalization with centromeres is a suitable method to judge sliding and end-on pulling, we expect that, when Kar3 is defective, lateral sliding (and conversion from sliding to end-on pulling) would become less frequent and, instead, end-on pulling would become more predominant, judged by this method. Using a kar3 temperature-sensitive mutant, we indeed confirmed this was the case (Supplementary Fig. S5).
Collectively, these results suggest that centromeres are transported poleward by both lateral sliding and end-on pulling during S phase. Note that, from Figure 7B, one may gain the impression that many cells use only one mode of transport, either lateral sliding or end-on pulling; however, many cells perhaps use both (lateral sliding and subsequently end-on pulling), especially if kinetochore transport events of distances <180 nm are also taken into account. Moreover, our new results, obtained in normal S phase, are consistent with the results recently obtained using the centromere reactivation system (Tanaka et al. 2007); namely, that (1) transport velocity is higher during end-on pulling than during lateral sliding, and (2) lateral sliding is converted to the end-on pulling, but the opposite conversion is rare.
Discussion
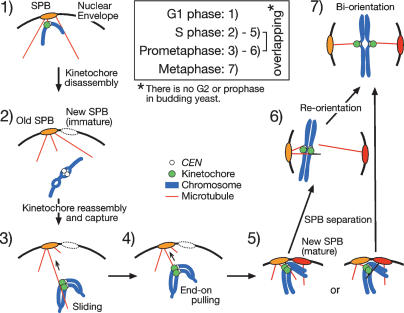
In the budding yeast S. cerevisiae, SPBs are embedded in the nuclear envelope and centromeres are tethered at SPBs via microtubules during G1 phase (Fig. 8, step 1; Supplementary Note 6; Guacci et al. 1997; Jin et al. 2000; Tanaka et al. 2002). It has been thought that centromeres might never detach from microtubules throughout the cell cycle. In contrast to this notion, we demonstrated here that centromeres become transiently (for 1–2 min) detached from microtubules in S phase, leading to centromere displacement away from a spindle pole (Fig. 8, step 2). Such centromere motion has been overlooked in the past, probably because it happens for such a short period. By setting a very short time interval of observation for live-cell imaging, we visualized this process for the first time. Centromere detachment and displacement are coincidental with, and actually dependent on centromere DNA replication in early S phase. In fact, if DNA replication was abolished by Cdc6 depletion, even though other cell cycle events were still ongoing, centromere detachment and displacement were very rarely observed.
Figure 8.
Summary of kinetochore–microtubule interaction from G1 to metaphase in S. cerevisiae. (Step 1) Kinetochores are attached to microtubules (perhaps to the ends of microtubules; see Supplementary Note 6) in G1. (Step 2) Kinetochores are disassembled upon centromere DNA replication, and centromeres are detached from microtubules and move away from a spindle pole. (Step 3) Kinetochores are reassembled, captured by the lateral surface of microtubules, and transported poleward by sliding along the microtubule surface. (Step 4) Kinetochores are tethered at the ends of microtubules and often further transported poleward as microtubules shrink. (Step 5) Both sister kinetochores interact with microtubules from either the same or different SPBs. (Steps 6, 7) SPBs separate at the end of S phase, and reorientation of kinetochore–microtubule attachment leads to sister kinetochore biorientation.
How does DNA replication cause centromere detachment from microtubules, but allows recapture soon afterward? Our data suggest that upon centromere DNA replication, kinetochores are disassembled, causing centromere detachment from microtubules, and they are subsequently reassembled, leading to centromere recapture by microtubules (Fig. 8, step 3; Supplementary Note 7). This indicates that, for a short period after CEN DNA replication, neither of the sister kinetochores is sufficiently intact to support microtubule attachment. In this regard, duplication of kinetochores following centromere DNA replication does not take place in a conservative manner (i.e., the old one from the previous cell cycle remains intact, while the new one is generated de novo in the present cycle) (Supplementary Notes 8, 9), in contrast to the duplication mechanism of SPBs (Adams and Kilmartin 2000; Pereira et al. 2001).
When centromeres are recaptured by microtubules, which SPB (i.e., new or old SPB) organizes these microtubules? The new SPB is formed de novo in the vicinity of the old SPB that has been inherited from the previous cell cycle (Adams and Kilmartin 2000; Pereira et al. 2001). SPB duplication proceeds during S phase (Lim et al. 1996), and therefore the new SPB might be too immature to generate microtubules when centromeres are ready for recapture by microtubules. This possibility was tested using an Ipl1 kinase mutant, in which the initial kinetochore–microtubule attachment state could be preserved due to a defect in subsequent reorientation (Tanaka et al. 2002). In this mutant, mono-orientation (where centromeres attach to microtubules only from one pole) was preferentially formed at the old SPB, suggesting that the new SPB was indeed immature and the old SPB usually organized microtubules for centromere recapture (Fig. 8, step 3). Supporting this notion, when centromeres were forced to replicate late in S phase so that more time was given for the new SPB to become mature, mono-orientation was formed equally at the new and old SPBs (Tanaka et al. 2002).
As discussed above, centromeres are initially captured by the lateral surface of microtubules (Fig. 8, step 3; K. Tanaka et al. 2005). Subsequently, centromeres are transported poleward in two distinct ways: lateral sliding and end-on pulling (Fig. 8, steps 3, 4; Tanaka et al. 2007). Sliding is often converted to end-on pulling, but the opposite conversion is rare. By these means, kinetochores reach the vicinity of a spindle pole, where the microtubule density becomes higher, thus allowing both sister kinetochores to interact with other microtubules more efficiently (Fig. 8, step 5). Meanwhile, the new SPB becomes mature enough to organize microtubules, and the Ipl1 kinase facilitates reorientation of kinetochore– microtubule attachment (Tanaka et al. 2002). Reorientation might involve transient detachment of centromeres from microtubules; nonetheless, this Ipl1-dependent detachment and DNA replication-dependent detachment of centromeres occur independently of each other. The Ipl1-dependent reorientation of kinetochore–microtubule attachment is regulated in a tension-dependent manner (Dewar et al. 2004). Thus, after SPBs separate and form a bipolar spindle at the end of S phase (Lim et al. 1996), this reorientation promotes sister kinetochore biorientation that generates tension at kinetochores (Fig. 8; steps 6, 7).
How are kinetochore–microtubule interactions similar and dissimilar between budding yeast and metazoan cells? In the open mitosis of metazoan cells, there is a large temporal gap (G2 phase) between DNA replication and the initial kinetochore–microtubule interaction, because MTOCs must wait for the nuclear envelope breakdown in order to organize microtubules for kinetochore capture (Sazer 2005). In contrast, in the closed mitosis of budding yeast, MTOCs are connected to kinetochores via microtubules throughout most of the cell cycle (Guacci et al. 1997; Jin et al. 2000; Winey and O’Toole 2001; Tanaka et al. 2002), becoming detached only for a short period in S phase. Considering that centromeres are recaptured by microtubules already during S phase, we propose that in budding yeast (1) there is no G2 phase (and no prophase) and (2) S phase and M phase (prometaphase) significantly overlap (Fig. 8, rectangle; Supplementary Note 10).
In spite of such difference, the mechanisms of kinetochore–microtubule interaction are remarkably similar in early mitosis (prometaphase and metaphase) (Lew and Burke 2003; T.U. Tanaka et al. 2005; Musacchio and Salmon 2007) between budding yeast and metazoan cells. In both, centromeres are initially captured by the lateral surface of microtubules and transported poleward along microtubules by minus-end-directed motors; subsequently, the Aurora B/Ipl1 kinase facilitates sister kinetochore biorientation, which is monitored by a conserved mechanism of spindle checkpoint; both biorientation and spindle-checkpoint mechanisms respond to tension applied on kinetochores, for which the conserved cohesin complex is required. Comparison of kinetochore–microtubule interaction between different organisms will uncover the evolution of regulatory mechanisms for this fundamental cellular process.
Materials and methods
Yeast genetics and molecular biology
The background of yeast strains (W303) and methods for yeast culture, α-factor treatment, and FACS DNA content analysis were as described previously (Amberg et al. 2005; Kitamura et al. 2006; Tanaka et al. 2007). Constructs of TetR-3CFP (Bressan et al. 2004), GFP-lacI (Straight et al. 1996), CEN5-tetOs (an array of 112×tetOs of 5.6 kb inserted at 1.4 kb left of CEN5) (Tanaka et al. 2000), CEN15-lacOs (an array of 256×lacOs of 10.1 kb inserted at 1.8 kb left of CEN15) (Goshima and Yanagida 2000), clb5 and clb6 deletion (Schwob and Nasmyth 1993), Pgal-CDC6 (Piatti et al. 1996), and GFP-TUB1 (Straight et al. 1997) were described previously. Mutant alleles ipl1-321 and kar3-64 were reported previously (Biggins et al. 1999; Cottingham et al. 1999). MTW1 and CTF19 were tagged with four tandem copies of GFP (4GFP), and ASK1 was tagged with 4GFP and with three tandem copies of CFP, at their C termini at their original gene loci by a one-step PCR method as described previously (Maekawa et al. 2003; K. Tanaka et al. 2005). GFP-TUB1 and YFP-TUB1 (pDH20, obtained from Yeast Resource Center) plasmids were integrated at auxotroph marker loci. Strains with the tagged genes grew normally at temperatures used in this study. Cells were cultured at 25°C in YP medium containing glucose, unless otherwise stated.
Microscopy
The procedures for time-lapse fluorescence microscopy were described previously (K. Tanaka et al. 2005; Tanaka et al. 2007). Time-lapse images were collected at 23°C (ambient temperature) unless otherwise stated. For image acquisition, we used a DeltaVision RT microscope (Applied Precision), a UPlanSApo 100× objective lens (Olympus; NA 1.40), SoftWoRx software (Applied Precision), and either a CoolSnap HQ (Photometrics) or Cascade II 512B (Roper Scientific) CCD camera. We acquired five to seven (0.7 μm apart) z-sections, which were subsequently deconvoluted, projected to two-dimensional images, and analyzed with SoftWoRx and Volocity (Improvision) software. CFP/GFP signals were discriminated from YFP using the JP3 filter set (Chroma). CFP signals were discriminated from either GFP or YFP using the JP4 filter set (Chroma). To collect GFP signals alone, the FITC filter (Chroma) was used.
Analyzing dynamics of kinetochores and microtubules
To evaluate the length of microtubules and position of centromeres, we took account of the distance along the Z-axis, as well as distance on a projected image. To avoid false judgement, detachment of GFP- and CFP-labeled CENs from GFP- or YFP-labeled microtubules was scored only when CEN signals did not overlap with microtubule signals for two or more consecutive time points. In the experiment shown in Figure 2B and Supplementary Figure S2, photobleaching of YFP-Tub1 signals was considerable during the 35–50 min after release from α-factor arrest; during this period, detachment was also scored when CEN–spindle pole distance was 700 nm or larger. To score the percentage of CEN detachment during a 5-min time window in Figures 2A, 3 and 5, the percentage of observed CEN detachment during the 4-min interval was multiplied by 1.25, and the last 1 min of each 5-min time window was used to change a microscopy field and to readjust focus. To measure cellular DNA content by FACS in Figures 2A and 3, samples were divided between microscopy observation and FACS analyses immediately after release from α-factor arrest; kinetics of bud progression was similar between the two samples. The start of CEN transport was defined as the time point from which CEN–spindle pole distance was first shortened for two or more consecutive time points after CEN and microtubule signals had overlapped again following CEN detachment. The end of CEN transport was defined as the time point at which CEN reached within 420 nm from a spindle pole. When microtubules was not visualized (Fig. 6; Supplementary Figs. S3–S5), the start of CEN transport was defined as the time point from which CEN–spindle pole distance was first shortened for two or more consecutive time points after CEN had moved away from a spindle pole (>700 nm), followed by shortening of this distance by 600 nm or more within 30 sec. The intensity of kinetochore component GFP signals at CFP-labeled CEN5 (Fig. 6; Supplementary Figs. S3, S4) was classified as follows: The integral GFP signal intensity (from each Voxel, volume pixel; with default setting of Volocity) colocalizing with the CEN5-CFP signal was scored as “−” for <35, “±” for 35∼100, and “+” for >100. Similarly, the presence of Ask1 signals at CEN15 (Fig. 7; or CEN5 in Supplementary Fig. S5) was scored when the integral Ask1-3CFP (or Ask1-4GFP) signal colocalizing with CEN15-GFP (or CEN5-CFP) signal was >60. Statistical analyses were carried out using the unpaired t-test (Figs. 1B, 7C), nonparametric tests (Mann-Whitney test or Kruskal-Wallis test) (Fig. 6; Supplementary Fig. S4) or χ2 test for trend (Supplementary Fig. S5), using the Prism (Graph pad) software, as explained in the figure legends. All P values are two-tailed. For more information, see Supplementary Note 11.
Acknowledgments
We thank L. Clayton, M.J.R Stark, J.-F. Maure, and members of the Tanaka laboratory for discussions and reading the manuscript; C. Allan and S. Swift for technical help for microscopy/computing; and K. Nasmyth, E. Schiebel, R. Tsien, K. Bloom, M.A. Hoyt, J.E. Haber, S. Biggins, M. Yanagida, A.W. Murray, A.F. Straight, J.-F. Maure, EUROSCARF, and the Yeast Resource Centre for reagents. This work was supported by Cancer Research UK, the Wellcome Trust, Human Frontier Science Program, Lister Research Institute Prize and Association for International Cancer Research. T.U.T. is a Senior Research Fellow of Cancer Research UK.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.449407
References
- Adams I.R., Kilmartin J.V., Kilmartin J.V. Spindle pole body duplication: A model for centrosome duplication? Trends Cell Biol. 2000;10:329–335. doi: 10.1016/s0962-8924(00)01798-0. [DOI] [PubMed] [Google Scholar]
- Amberg D.C., Burke D.J., Strathern J.N., Burke D.J., Strathern J.N., Strathern J.N. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Sassoon I., Hyman A.A., Murray A.W., Hyman A.A., Murray A.W., Murray A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Tanaka T.U., Tanaka T.U. The chromosome cycle: Coordinating replication and segregation. Second in the cycles review series. EMBO Rep. 2005;6:1028–1034. doi: 10.1038/sj.embor.7400557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Bressan D.A., Vazquez J., Haber J.E., Vazquez J., Haber J.E., Haber J.E. Mating type-dependent constraints on the mobility of the left arm of yeast chromosome III. J. Cell Biol. 2004;164:361–371. doi: 10.1083/jcb.200311063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F.R., Gheber L., Miller D.L., Hoyt M.A., Gheber L., Miller D.L., Hoyt M.A., Miller D.L., Hoyt M.A., Hoyt M.A. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T.U., Tanaka K., Nasmyth K., Tanaka T.U., Nasmyth K., Tanaka T.U., Tanaka T.U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Goshima G., Yanagida M., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D., Hogan E., Koshland D., Koshland D. Centromere position in budding yeast: Evidence for anaphase A. Mol. Biol. Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P., Laroche T., Shimada K., Furrer P., Gasser S.M., Laroche T., Shimada K., Furrer P., Gasser S.M., Shimada K., Furrer P., Gasser S.M., Furrer P., Gasser S.M., Gasser S.M. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Hill A., Bloom K., Bloom K. Genetic manipulation of centromere function. Mol. Cell. Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Winey M., Winey M. The budding yeast spindle pole body: Structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jin Q.W., Fuchs J., Loidl J., Fuchs J., Loidl J., Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 2000;113:1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Kitamura E., Blow J.J., Tanaka T.U., Blow J.J., Tanaka T.U., Tanaka T.U. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Sandall S., Desai A., Sandall S., Desai A., Desai A. Kinetochore–spindle microtubule interactions during mitosis. Curr. Opin. Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., Burke D.J., Burke D.J. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- Lim H.H., Goh P.Y., Surana U., Goh P.Y., Surana U., Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol. Cell. Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui T., Knop M., Schiebel E., Usui T., Knop M., Schiebel E., Knop M., Schiebel E., Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Deluca J., Salmon E.D., Earnshaw W.C., Deluca J., Salmon E.D., Earnshaw W.C., Salmon E.D., Earnshaw W.C., Earnshaw W.C. The dynamic kinetochore–microtubule interface. J. Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- McAinsh A.D., Tytell J.D., Sorger P.K., Tytell J.D., Sorger P.K., Sorger P.K. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- McCarroll R.M., Fangman W.L., Fangman W.L. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J., Stemmann O., Rank S., Lechner J., Rank S., Lechner J., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes & Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Yeh E., Gardner M., Odde D., Salmon E.D., Bloom K., Yeh E., Gardner M., Odde D., Salmon E.D., Bloom K., Gardner M., Odde D., Salmon E.D., Bloom K., Odde D., Salmon E.D., Bloom K., Salmon E.D., Bloom K., Bloom K. Stable kinetochore–microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka T.U., Nasmyth K., Schiebel E., Tanaka T.U., Nasmyth K., Schiebel E., Nasmyth K., Schiebel E., Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p–Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer C., Nasmyth K., Lengauer C., Nasmyth K., Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm T., Cocker J.H., Diffley J.F., Nasmyth K., Bohm T., Cocker J.H., Diffley J.F., Nasmyth K., Cocker J.H., Diffley J.F., Nasmyth K., Diffley J.F., Nasmyth K., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Davis R.W., Brewer B.J., Fangman W.L., Brewer B.J., Fangman W.L., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Sazer S. Nuclear envelope: Nuclear pore complexity. Curr. Biol. 2005;15:R23–R26. doi: 10.1016/j.cub.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Schwob E., Nasmyth K., Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes & Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Severin F., Hyman A.A., Piatti S., Hyman A.A., Piatti S., Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Centrosome duplication. A centriolar pas de deux. Cell. 2001;105:417–420. doi: 10.1016/s0092-8674(01)00366-x. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W., Belmont A.S., Robinett C.C., Murray A.W., Robinett C.C., Murray A.W., Murray A.W. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Marshall W.F., Sedat J.W., Murray A.W., Marshall W.F., Sedat J.W., Murray A.W., Sedat J.W., Murray A.W., Murray A.W. Mitosis in living budding yeast: Anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U. Chromosome bi-orientation on the mitotic spindle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:581–589. doi: 10.1098/rstb.2004.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Knapp D., Nasmyth K., Knapp D., Nasmyth K., Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K., Fuchs J., Loidl J., Nasmyth K., Loidl J., Nasmyth K., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Galova M., Schiebel E., Stark M.J., Nasmyth K., Schiebel E., Stark M.J., Nasmyth K., Stark M.J., Nasmyth K., Nasmyth K. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U., Mukae N., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U., James E.K., Prescott A.R., Antony C., Tanaka T.U., Prescott A.R., Antony C., Tanaka T.U., Antony C., Tanaka T.U., Tanaka T.U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Stark M.J., Tanaka K., Stark M.J., Tanaka K., Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nat. Rev. Mol. Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T.U., Kitamura E., Kitamura Y., Tanaka T.U., Kitamura Y., Tanaka T.U., Tanaka T.U. Molecular mechanisms of microtubule-dependent kinetochore trasport towards spindle poles. J. Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., O’Toole E.T., O’Toole E.T. The spindle cycle in budding yeast. Nat. Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]