Abstract
O6-Methylguanine (MeG) is a highly cytotoxic DNA modification generated by SN1-type methylating agents. Despite numerous studies implicating DNA replication, mismatch repair (MMR), and homologous recombination (HR) in MeG toxicity, its mode of action has remained elusive. We studied the molecular transactions in the DNA of yeast and mammalian cells treated with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). Although replication fork progression was unaffected in the first cell cycle after treatment, electron microscopic analysis revealed an accumulation of MeG- and MMR-dependent single-stranded DNA (ssDNA) gaps in newly replicated DNA. Progression into the second cell cycle required HR, while the following G2 arrest required the continued presence of MeG. Yeast cells overcame this block, while mammalian cells generally failed to recover, and those that did contained multiple sister chromatid exchanges. Notably, the arrest could be abolished by removal of MeG after the first S phase. These new data provide compelling support for the hypothesis that MMR attempts to correct MeG/C or MeG/T mispairs arising during replication. Due to the persistence of MeG in the exposed template strand, repair synthesis cannot take place, which leaves single-stranded gaps behind the replication fork. During the subsequent S phase, these gaps cause replication fork collapse and elicit recombination and cell cycle arrest.
[Keywords: DNA repair, DNA replication, methylating agents, mismatch repair, replication intermediates]
The SN1-type chemotherapeutic agents procarbazine, dacarbazine, temozolomide, and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) methylate the N7 and O6 atoms of guanine, the N1 and N3 atoms of adenine, and the N3 atom of cytosine. The N-methylated purines, which account for ∼80% of the modifications, are repaired rapidly and efficiently by base excision repair (BER), causing no cytotoxicity at clinically relevant doses. In contrast, the low amounts (∼8%) of O6-methylguanine (MeG) are sufficient to cause cell death. Under normal circumstances, the methyl group is removed from the 6 position of guanine by methylguanine methyl transferase (MGMT), a repair protein that is targeted for degradation after transferring the methyl group from the guanine to a cysteine residue in its active site (Pieper 1997). Correspondingly, cells expressing high levels of MGMT are resistant to methylating agents, while prolonged treatment of cells expressing low amounts of MGMT leads to enzyme depletion and drug sensitization. This property of MGMT is currently being exploited in chemotherapy, where pretreatment with the MGMT competitive inhibitor O6-benzylguanine (BG) is used to deplete the levels of this repair protein and thus sensitize cells to killing by SN1-type methylating agents (Dolan et al. 1998).
In the absence of MGMT, MeG persists in DNA and brings about cell cycle arrest and cell death in a mismatch repair (MMR)-dependent manner. How these events are triggered is currently the subject of extensive discussions. Because DNA replication is absolutely required for MNNG-mediated cytotoxicity (Kaina 2004), it is believed that the primary lesion activating the sequence of events that culminate in cell death is the formation of MeG/T and MeG/C mispairs generated by replicating DNA polymerases when they encounter MeG in the template strand. As both these mispairs are bound by the mismatch recognition factor MutSα (Duckett et al. 1996; Berardini et al. 2000), it has been proposed that this binding activates the apoptotic signaling cascade (Fishel 1998; Yang et al. 2004; Yoshioka et al. 2006). An alternative hypothesis posits that cell death is caused by the unsuccessful processing of the MeG-containing base pairs by the MMR system, which activates a cell cycle arrest (Stojic et al. 2004) from which most cells fail to recover. This latter hypothesis is supported by the finding that the arrest is activated only in the second S phase after MNNG treatment (Zhukovskaya et al. 1994; Stojic et al. 2004), whereas the direct signaling hypothesis would predict that the cells should arrest after the first S phase, given that MeG-containing mispairs are formed already during the first replication.
In eukaryotic cells, MMR is tightly linked with replication through PCNA, which interacts directly with MutSα (Clark et al. 2000; Kleczkowska et al. 2001) and thus provides MMR with ready access to mismatches generated by the polymerases. It might therefore be anticipated that when MMR attempts to process a MeG-containing mispair, the replicating polymerase might be retarded, because the modified guanine in the template strand does not have a perfect Watson-Crick partner, and thus that the MMR process cannot be completed. Should the presence of MeG in the template DNA fail to cause a significant DNA synthesis delay, it would imply that the processing of this lesion is decoupled from replication. The first goal of this study was to address this issue.
That loss of homologous recombination (HR) sensitizes MMR-proficient yeast and mammalian cells to killing by MNNG (Cejka et al. 2005; Tsaryk et al. 2006) provided further support for the processing hypothesis, suggesting that intermediates arising during the futile attempts of the MMR machinery to correct MeG-containing mispairs are channeled into HR. However, these studies failed to reveal the possible structure(s) of the HR intermediates. The second goal of this study was therefore to gain novel insights into the MMR-dependent transactions in the DNA of MNNG-treated mammalian and yeast cells. We also wanted to identify the processes that allow the treated cells to bypass MeG during the first S phase and to undergo cell division with damaged DNA.
Results
MNNG-induced arrest in the second cell cycle is dependent solely on MeG
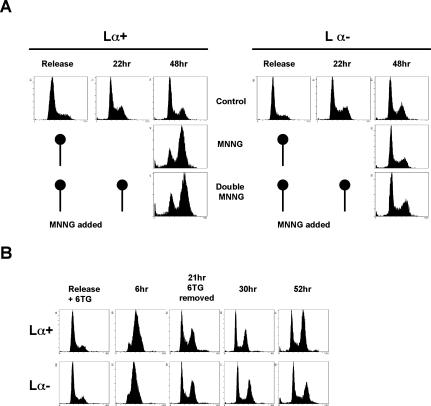
Exposure of mammalian cells to MNNG brings about a MMR-dependent cell cycle arrest in the second G2 phase after treatment (Stojic et al. 2004). However, the MeG/T and MeG/C mispairs, which are believed to activate MMR, arise already in the first S phase, so why do they fail to trigger the cell cycle arrest then? How does the DNA of newly treated cells differ from that of cells that have undergone one division? As mentioned above, MeG is only a minor product of MNNG treatment. This chemical generates predominantly N7-methyguanine and N3-methyladenine, which are not substrates for MMR, but which are rapidly and efficiently repaired by BER (Wyatt and Pittman 2006). The latter modifications will thus have been removed from DNA prior to the onset of the second S phase, and only MeG will persist. We therefore wanted to test whether it was the processing of the N-methylated purines during the first cell cycle that allowed the cells to progress to the next one. In this scenario, a second treatment with MNNG after the cell division should carry the cells also across the second mitotic boundary, and the activation of the cell cycle arrest should be delayed until the third cycle. In order to test this possibility, we used 293T Lα cells, which are MGMT- and MLH1-deficient, but carry a stably integrated hMLH1 cDNA minigene controlled by the TetOffTM expression system (Cejka et al. 2003). In the absence of doxycycline (Dox), these cells, referred to as 293T Lα+, express hMLH1 and are MMR-proficient and MNNG-sensitive. In contrast, when the same cells are grown in the presence of 50 ng/mL Dox (293T Lα− cells), they shut off hMLH1 expression and become MMR-deficient and resistant to MNNG. We synchronized the 293T Lα cells with 2 mM hydroxyurea (HU) for 16 h (Fig. 1A) and released them into fresh medium without (Control) or with 0.2 μM MNNG. The cells were then treated with 0.2 μM MNNG a second time, 22 h after release, when the majority were in the G1 phase of the second cell cycle (Double MNNG). As the cells treated at the point of release from the HU block and those treated with MNNG twice arrested at the same time point, we concluded that processing of lesions other than MeG does not promote cell cycle progression after MNNG treatment.
Figure 1.
MNNG cell cycle arrest depends on MeG alone and not on other types of methylation damage. (A) Lesions other than MeG do not mediate progression from the second to the third cell cycle. After release from HU synchronization, 293T Lα cells were treated with MNNG and allowed to progress to the following G1 phase, where they were treated with MNNG again in order to create lesions of similar type and quantity as during the first treatment. As can be seen, the presence of these modifications did not influence MMR-dependent arrest, as the cells still arrested in the second G2/M phase after the first treatment. (B) MeTG causes an MMR-dependent arrest identical to that induced by MNNG, suggesting that the cell cycle arrest is activated by methylation of O6 of guanine or S6 of thioguanine, rather than by other types of damage.
To further confirm the above hypothesis, we treated the cells with 6-thioguanine (6TG), which is incorporated into DNA and converted to MeTG, an analog of MeG (Karran 2006). The DNA of cells treated with TG thus does not contain modified nucleotides other than MeTG. As shown in Figure 1B, treatment of synchronized 293T Lα cells with 3 μM 6TG for 21 h brought about a MMR-dependent arrest two cell cycles after treatment (52 h after release), similar to that induced by MNNG (Stojic et al. 2004). Taken together, the above data show that the MNNG-induced second cell cycle arrest is triggered by the processing of a single lesion, namely, MeG.
MMR-dependent processing of MeG does not affect replication fork progression, but leads to a reduced replication rate in the second cell cycle
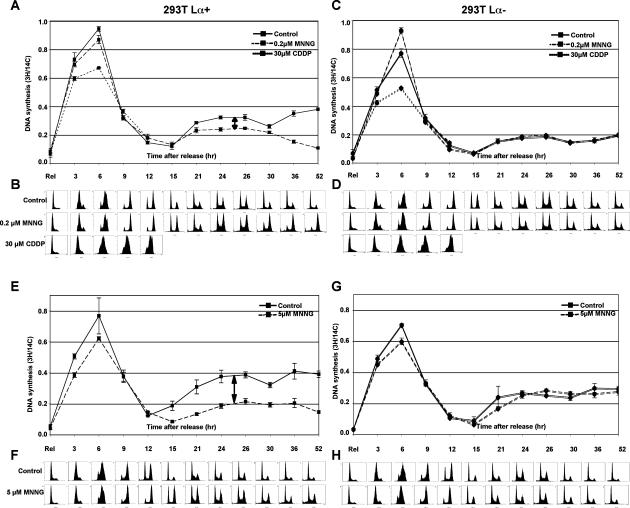
Our previous experiments (Stojic et al. 2004) suggested that progression of the cells through the first S phase was largely unaffected by MNNG treatment, at least as judged by flow cytometric analysis (see also Fig. 7A, below). However, we wanted to study the replication rates of treated and untreated cells in greater detail. Using 293T Lα cells, we measured the effect of two different concentrations of MNNG on the incorporation of radioactive nucleotides into genomic DNA. As a positive control, we used cisplatin (CDDP), a cross-linking agent known to interfere with replication fork progression (Henry-Mowatt et al. 2003). As shown in Figure 2A,C,E,G, DNA synthesis levels were highest during the first S phase and were similar in both 0.2 μM MNNG-treated and control samples, while the replication rates of cisplatin-treated cells were substantially lower in both 293T Lα+ and 293T Lα− cells (Fig. 2A,C). The slight decrease in incorporation rates observed after 5 μM MNNG treatment was largely MMR independent, as it was similar in both 293T Lα+ and 293T Lα− cells (Fig. 2E,G). The above cell populations were followed in parallel by flow cytometry, where we observed a slight delay in the exit of the MMR-proficient cells from the first G2 phase (12 and 15 h) after MNNG treatment (Fig. 2, cf. B,F and D,H). As anticipated, at time points corresponding to the second S phase (21–30 h), a new incorporation rate increase was detected (Fig. 2A,C,E,G), but the peaks were lower and broader than in the first S phase, due to a partial loss of synchronization of the cell population (Fig. 2B,D,F,H). At these time points, the MMR-proficient 293T Lα+ cells displayed a concentration-dependent decrease in incorporation rates compared with control cells (Fig. 2A,E, two-way arrows). The observed difference became more pronounced at later time points, when the MMR-proficient cells became arrested while the control cells continued to proliferate. No difference was observed in the MMR-deficient 293T Lα− cells (Fig. 2C,G). The MMR-dependent decrease in DNA synthesis rates may appear small, but it gains in significance when one takes into account that, at these time points, the proportion of S-phase cells in MNNG-treated cells was higher than in the untreated cells (Fig. 2B,F). These findings thus indicate that while MNNG treatment does not affect DNA synthesis during the first S phase, a significant fraction of forks in the second S phase of MMR-proficient cells are not actively replicating.
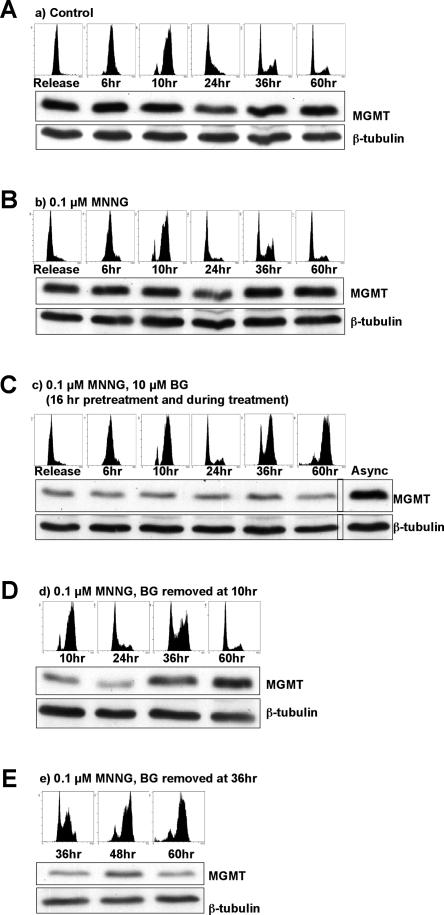
Figure 7.
MNNG-activated cell cycle arrest requires the persistence of MeG in DNA through two cell cycles. Cells were synchronized with 2 mM HU and released into fresh medium (A) or medium containing 0.1 μM MNNG (B–E). (C) Cells incubated with BG arrested at G2/M two cell cycles after release from HU. (D) When BG was removed 10 h after release (when cells passed the first S phase), the cells continued to proliferate without arresting. (E) In contrast, if BG was removed at the point when the checkpoint was already activated, the cells remained arrested.
Figure 2.
MMR-dependent processing of MeG does not affect replication fork progression but leads to a reduced replication rate in the second cell cycle. (A,C,E,G) Incorporation rate of tritiated thymidine was measured in 293T Lα cells upon release from HU synchronization after treatment with two different concentrations of MNNG. (B,D,F,H) In parallel, cell cycle progression was followed by DNA content analysis. The incorporation rates during the first cell cycle were similar, indicating that processing of the MeG-containing mispairs by MMR does not affect progression of the replication forks. However, replication efficiency was dramatically affected during the second cell cycle (arrows) in a dose-dependent manner, and only in MMR-proficient cells. All experiments were repeated at least three times in triplicate. The figure shows representative profiles of one single experiment.
Because of its smaller genome size and known replication origins, we wanted to study the effect of MNNG treatment on replication fork progression in Saccharomyces cerevisiae. Despite the evolutionary conservation of MMR, yeast cells are, unlike their human counterparts, highly resistant to MNNG treatment. However, their MMR-dependent MNNG sensitivity becomes apparent in recombination (rad52) and methylguanine methyltransferase (mgt1) deficient background (Cejka et al. 2005). Neutral–neutral two-dimensional DNA electrophoresis (2D gels) (Lopes et al. 2006) failed to reveal evidence of replication fork stalling or slow-down after MNNG treatment in cells either sensitive (mgt1 rad52) or resistant (mgt1 rad52 msh2) to MNNG (Supplementary Fig. 1). We therefore carried out an electron microscopic (EM) analysis of structures of replication intermediates (RIs) isolated from both yeast and mammalian cells synchronized with α-factor or HU, respectively, treated with MNNG, and allowed to progress to the first S phase (30 min for yeast, 4 h for human cells). We could show previously that yeast replication forks contain short (<400-nucleotide [nt]) single-stranded DNA (ssDNA) regions on one of the nascent strands emanating from the fork, while extended ssDNA regions (up to 3 kb) can be detected at 30%–50% of the replication forks after treatments that affect replication fork progression (Lopes et al. 2006). In the untreated mgt1 rad52 yeast strain and in 293T Lα cells, the proportion of forks with ssDNA regions >400 nt was ∼5% (Supplementary Fig. 2). It increased only slightly upon MNNG treatment (3 μM) and appeared to be MMR independent (mgt1 rad52 vs. mgt1 rad52 msh2) and MeG independent (mgt1 rad52 vs. MGT1 rad52). This suggests that the occasional extended ssDNA regions at the fork are linked to processing of lesions other than MeG. Thus, our data suggest that replication fork progression is largely unaffected both in yeast and human cells during the first replication after MNNG treatment.
MMR-dependent ssDNA gaps accumulate behind replication forks encountering MeG in MNNG-treated yeast and mammalian cells
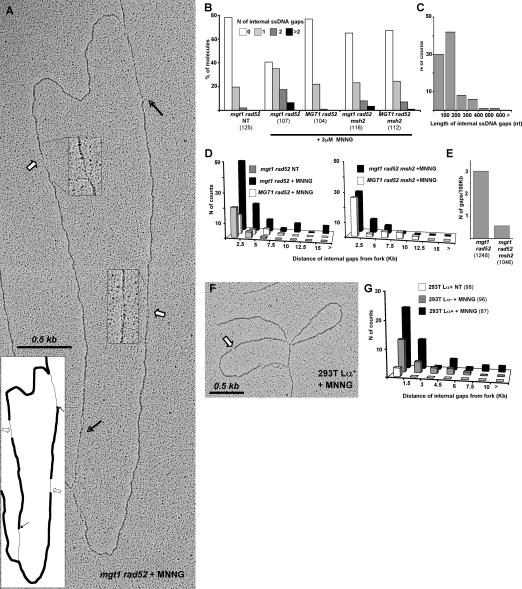
Despite the apparent lack of a MeG-dependent effect on fork progression and structure, the EM analysis of the replicated duplexes behind the fork was informative. Untreated mgt1 rad52 cells contained an elevated number of RIs with ssDNA regions in replicated duplexes (internal gaps) when compared with untreated wild-type cells, which was seen also in other DNA-repair-deficient yeast strains. This difference is likely linked with the inability of the former cells to process endogenous DNA damage (Lopes et al. 2006). In MNNG-treated mgt1 rad52 cells, >50% of the RIs contained internal gaps (Fig. 3A,B), of which ∼20% contained multiple gaps. This increase was absolutely dependent on MeG formation, as RIs from MGT1 rad52 cells, which can readily remove methyl groups from the O6 position of guanines, did not show elevated levels of internal gaps when compared with the untreated control. A similarly low number of ssDNA gaps was observed in RIs of MNNG-treated mgt1 rad52 msh2 and MGT1 rad52 msh2 strains. This showed that formation of the internal gaps is dependent on functional MMR (Fig. 3B).
Figure 3.
MMR- and MeG-dependent ssDNA gaps accumulate behind yeast and mammalian replication forks encountering MNNG-damaged templates. (A) Electron micrograph of a representative RI isolated from mgt1 rad52 cells treated with 3 μM MNNG and cross-linked in vivo with psoralen 30 min after release from G1. White arrows and magnified inserts indicate ssDNA regions along the replicated strands (internal gaps), and black arrows indicate the transition from double-stranded DNA (dsDNA) to ssDNA at the replication fork. (B) Distribution of ssDNA gaps in yeast strains treated with 3 μM MNNG. The increased number of gaps was dependent on the presence of MGT1 and functional MMR. The total number of molecules analyzed is shown in parentheses. (C) Lengths of internal gaps in the mgt1 rad52 strain. The majority of the gaps were <200 nt, independently of genetic background (data not shown). (D) Distribution of internal gaps relative to the replication forks. The population of molecules analyzed was the same as in Figure 2B. The increase in the number of gaps far from the fork was dependent on MGT1 and MMR. (E) Number of internal gaps in fully replicated DNA isolated from cells 90 min after release from G1. MMR-proficient cells displayed an increase in the number of gaps along replicated duplexes after MNNG treatment. The total length (in kilobases) of linear replicated DNA analyzed is shown in parentheses. (F) Electron micrograph of a representative RI isolated from 293T Lα+ cells treated with 5 μM MNNG and cross-linked in vivo with psoralen 4 h after release from G1. (G) Distribution of internal gaps relative to replication forks in 293T Lα cells. MNNG treatment brought about an increase in the number of gaps along the replicated duplexes, with 293T Lα+ cells displaying a significant proportion of gaps far from the fork. The total number of molecules analyzed is shown in parentheses. All experiments were carried out at least twice, and the observed differences could be shown to be reproducible. Due to subtle, uncontrollable variations in sample preparation, results from independent experiments cannot be directly averaged. Hence, the figure shows representative graphs from one single experiment.
We then wanted to find out whether functional MMR affects the length and distribution of the gaps. In untreated cells, they were predominantly found within 2.5 kb behind the fork, implying that such gaps are promptly repaired as the fork progresses. In contrast, in mgt1 rad52 cells treated with 3 μM (Fig. 3C), as well as in other genetic backgrounds (data not shown), the gaps were generally <200 nt and many were found >7.5 kb from the fork. This phenotype was entirely dependent on MeG, as the distant gaps were absent from RIs of MNNG-treated MGT1 rad52 cells. Similarly, in RIs of MMR-deficient mgt1 rad52 msh2 cells, the proportion of gaps close to the fork was comparable with that seen in untreated cells, but the distant gaps were largely absent. As anticipated, the effect of MGT1 in the absence of MMR was substantially lower than in MMR-proficient cells.
The larger number of gaps in close proximity to the fork in MNNG-treated mgt1 rad52 cells suggests that the gaps were generated during replication, but that they did not affect fork progression (Supplementary Figs. 1, 2). To assess the persistence of the gaps, we analyzed DNA of cells allowed to progress for 90 min after release from α-factor and MNNG treatment. At this time point, the majority of cells were in G2 (Fig. 5D, below; data not shown) and their DNA was fully replicated. As shown in Figure 3E, DNA from mgt1 rad52 cells contained approximately five times more ssDNA gaps than that isolated from mgt1 rad52 msh2 cells, which showed that the persistence of the gaps in replicated duplexes is linked with functional MMR.
Figure 5.
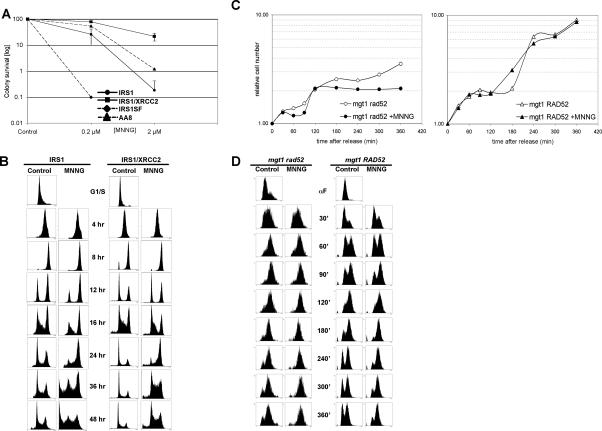
Recombination is required for transition from the first into the second cell cycle after MNNG treatment in both mammalian and yeast cells. (A) Colony survival assay of HR-deficient cell lines after MNNG treatment. Cell lines deficient in the RAD51 paralogs XRCC2 (IRS1) and XRCC3 (IRS1SF) are hypersensitive to MNNG compared with their recombination-proficient counterparts. (B) HR-deficient IRS1cells arrested in the first G2 phase after MNNG treatment (12–16 h), whereas the HR-proficient IRS1/XRCC2 cells went through the first cell cycle and arrested in the second G2 after treatment. (C) Growth curves of HR-deficient mgt1 rad52 and HR-proficient mgt1 RAD52 yeast cells. The growth of mgt1 RAD52 cells was unaffected by MNNG treatment, whereas mgt1 rad52 cells stopped growing after one cell cycle. (D) The mgt1 rad52 strain arrests in the first G2/M after MNNG treatment. Although the mgt1 RAD52 cells continued to grow after treatment, they spent longer in G2.
This analysis was extended to RIs isolated from human 293T Lα cells 4 h after release from HU arrest, when the cells were in S phase. The contribution of MMR to the number and distribution of the ssDNA gaps observed in yeast cells was seen also in human cells, even though it was less prominent (Fig. 3F). Upon treatment with 5 μM MNNG, the number of gaps in nascent DNA increased significantly in both MMR-proficient (293T Lα+) and -deficient (293T Lα−) cells, but their number and persistence farther away from the fork was substantially higher in the former (Fig. 3G).
Taken together, these data indicate that MMR-independent gaps, which arise most likely through processing of lesions other than MeG, are rapidly repaired behind the progressing replication fork. In contrast, gaps arising through MMR-dependent processing of MeG remain in the DNA even after the completion of S phase.
MNNG treatment brings about retention of PCNA in chromatin
The above experiments, together with in vitro evidence from the Modrich laboratory (York and Modrich 2006), support the hypothesis that MNNG-induced damage is actively processed. In an attempt to visualize this processing, we followed the distribution of PCNA in the treated cells. In actively replicating cells, PCNA in replication factories is chromatin bound and appears as bright foci. Cells in other phases of the cell cycle contain soluble PCNA, as witnessed by a lack of signal upon staining with PCNA antibodies (Fig. 4A). We synchronized MGMT-deficient, MMR-proficient HeLa mex− cells with double-thymidine block and released them into fresh medium with or without 0.4 μM MNNG. At the indicated time points, the cells were incubated in permeabilization buffer (see Materials and Methods) prior to fixation in ice-cold methanol. This procedure, developed for the study of MRE11 (Mirzoeva and Petrini 2001), permits the extraction of soluble proteins that are not bound to chromatin.
Figure 4.
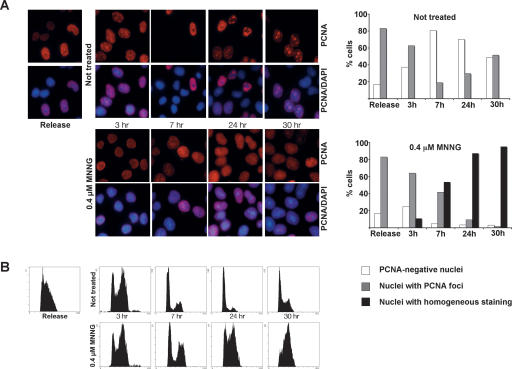
MNNG treatment brings about a retention of PCNA in chromatin. (A) Cells synchronized in G1/S were released and either left untreated or treated with 0.4 μM MNNG. After permeabilization prior to fixation, only chromatin-bound PCNA remained in the cells. (B) Although both control and treated cells progressed through the cell cycle, PCNA was retained in the chromatin only in the MNNG-treated cells. The right panel in A shows a graphic representation of the different types of PCNA staining.
The cells were MNNG-treated at the point of release from the double-thymidine block, when the majority were in G1/early S phase, and their progress through the cell cycle was monitored by flow cytometry. Three hours later, both control and MNNG-treated cells were in late S phase, where 60%–70% of the cells displayed prominent PCNA staining, indicative of chromatin-bound protein engaged in DNA replication. Seven hours after release, the majority of both control and treated cells were in the G1 phase of the second cell cycle. In the untreated sample, only a minor fraction of the cells was PCNA-positive. In contrast, ∼90% of the cells treated with MNNG displayed strong PCNA staining, even though cell cycle analysis indicated that the majority was in G1. In most cells, the pattern of PCNA staining was also somewhat different from that seen in the unperturbed S phase. The MNNG-dependent staining was not as intense and lacked the focal pattern characteristic of replicating cells; rather, the staining was relatively homogeneous. At the later time points (24 and 30 h after release), the untreated cells reverted to asynchronous profiles, and the PCNA staining revealed a mixed population of cells with and without foci, indicative of cells in different cell cycle phases. In contrast, MNNG-treated cells were accumulating in the second S phase, suggesting that their DNA replication had been impaired. The PCNA staining of the treated cells was also substantially different from that of the untreated cells. Specifically, we observed a fairly intense homogeneous staining in >90% of the cells, and although the cells at these time points (24 and 30 h) were in the phase of the cell cycle that resembled the point of release as judged by cell cycle analysis, the staining pattern of chromatin-bound PCNA was remarkably different. Particularly striking was the lack of foci, characteristic of S phase in untreated cells. Thus, although progression through the first cell cycle was not significantly affected by MNNG treatment (Fig. 4B, Release to 7 h), the persistence of chromatin-bound PCNA is suggestive of DNA processing that is uncoupled from replication. This processing continued into the second cell cycle, as witnessed by the retention of diffuse PCNA staining in chromatin.
Transition of MNNG-treated cells from the first into the second cell cycle requires recombination
The persistence of chromatin-bound PCNA staining in MNNG-treated cells suggested that replication-independent DNA processing was occurring already during the first cell cycle after treatment. That MNNG treatment brings about activation of HR in bacteria (Nowosielska et al. 2006), yeast (Cejka et al. 2005), and mammalian cells (Zhang et al. 2000; Tsaryk et al. 2006) has been well documented. We wanted to test whether recombination proficiency affects the sensitivity of cells to this agent. To this end, we studied the MNNG response of two matched pairs of cell lines: the XRCC2-deficient Irs1 cells and their derivative in which the HR defect was complemented with XRCC2 cDNA, and the XRCC3-deficient Irs1SF cells and their parental, HR-proficient line AA8. Irs1 cells fail to form RAD51 foci in response to DNA damage and are hypersensitive to a variety of DNA damaging agents such as the cross-linking agent Mitomycin C (Jones et al. 1987). Expression of human XRCC2 cDNA in these cells restores DNA damage resistance and RAD51 focus formation (O’Regan et al. 2001). Irs1SF cells are similarly hypersensitive to DNA damaging agents.
As shown in Figure 5A, cell survival after MNNG treatment was highly dependent on functional HR, as shown recently also for HR-deficient mouse embryonic fibroblasts (MEFs) (Tsaryk et al. 2006). The Irs1 and Irs1SF lines were substantially more sensitive to MNNG than their recombination-proficient counterparts. We wanted to learn how HR deficiency affected treatment-dependent cell cycle distribution and arrest. As shown in Supplementary Figure 3, the HR-proficient Irs1/XRCC2 and AA8 cells displayed a typical profile of MNNG-treated MMR-proficient cells. They required two DNA replication cycles to activate the checkpoint and arrested in G2/M. In contrast, the HR-deficient Irs1 and Irs1SF lines accumulated in G2/M phase already 16 h after MNNG treatment, a time period during which the cells replicated their DNA only once.
To follow the MMR-dependent cell cycle arrest in recombination-proficient and -deficient cells in more detail, we synchronized the Irs1 and Irs1/XRCC2 cells in G1/S and released them into medium with or without MNNG (Fig. 5B). Control synchronizations showed that both HR-proficient and -deficient cells readily recovered from the HU-induced arrest. Progression of the Irs1/XRCC2 cells followed an already known pattern of MMR-proficient cells, where they clearly entered the second cell cycle after treatment as indicated at 12 and 16 h after release by an increase in the G1 fraction. In contrast, the Irs1 cells were still in G2 at these time points, with only a very small proportion entering the second cycle. The difference was also apparent at later time points, where many more Irs1 cells were detected in the sub-G1 region of the histogram, which is indicative of dead cells.
To assess the contribution of HR to cell cycle progression of yeast cells, the recombination-proficient mgt1 RAD52 and recombination-impaired mgt1 rad52 strains were synchronized by α-factor and released, and the cells were counted for the following 360 min, which corresponded to approximately three cell cycles. The recombination-proficient mgt1 RAD52 cells were not affected by MNNG treatment and continued to double their number similarly to the untreated control. In contrast, the recombination-deficient mgt1 rad52 cells doubled only once in the presence of MNNG, even when the substantially slower growth of these cells, linked most likely with their reduced ability to repair endogenous damage, was taken into account (Fig. 5C). Unlike their mammalian counterparts, the recombination-proficient yeast cells did not display an MMR-dependent arrest after MNNG treatment, and their growth appeared to be unaffected. Flow cytometric analysis revealed that the MNNG-treated mgt1 rad52 cells arrested in the first G2/M, whereas the untreated cells re-entered the cell cycle (Fig. 5D, 180–360 min). This was confirmed by growth analysis, which showed that the treated cells stopped dividing (Fig. 5C). Surprisingly, the fraction of mgt1 RAD52 cells in G2/M increased, even though this strain did not show any growth delay (Fig. 5D, 180–360 min).
Recombination has different roles in the first and second cell cycle after MNNG treatment
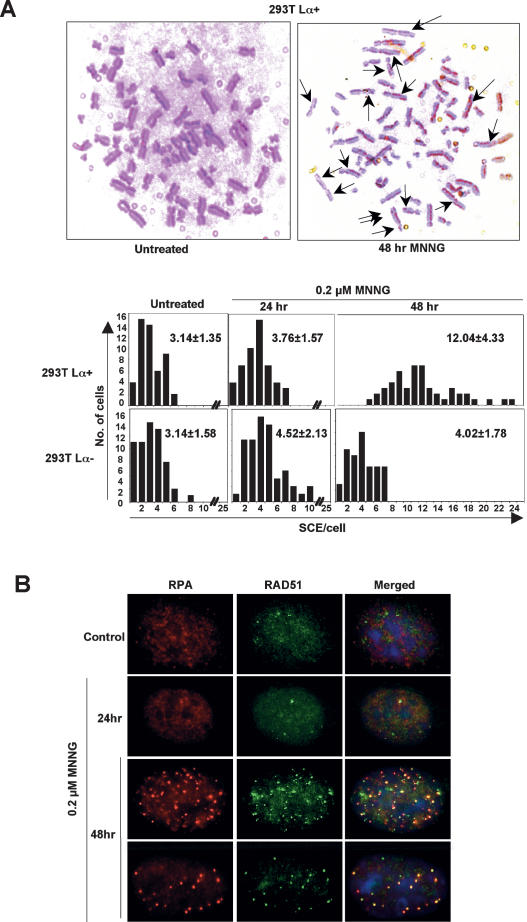
That MNNG treatment triggers HR has been known for some time. In mammalian cells it causes an elevated frequency of sister chromatid exchanges (SCEs), which have been attributed to MeG (Kaina and Aurich 1985; Kaina 2004). In order to test whether SCE formation is also dependent on MMR, we examined mitotic chromosomes of both MMR-proficient and -deficient 293TLα cells after MNNG treatment. SCE formation was monitored after 24 h, when the cells have undergone only one replication in the presence of MeG, or after 48 h. As shown in Figure 6A, elevated SCE levels were observed only in 293TLα+ cells and only 48 h after MNNG treatment; the number of SCEs per cell rose from an average of 3.14 to about four times as much, with many cells having as many as 20 SCEs.
Figure 6.
Recombination mechanisms in the first and second cell cycles after MNNG treatment differ. (A) Representative images of metaphase spreads of 293T Lα+ cells stained to differentiate sister chromatids of individual chromosomes. Only MMR-proficient cells displayed elevated levels of SCEs, and only 48 h after MNNG treatment. (B) Coimmunostaining of MMR-proficient HeLa cells showing colocalization of RPA and RAD51 foci after 0.2 μM MNNG treatment.
Another marker of HR activation in living cells is formation of foci containing the ssDNA-binding proteins RAD51 or RPA (Raderschall et al. 1999). In asynchronous 293TLα+ cells, RAD51 foci became apparent 24 h after MNNG treatment and were most numerous 1 d later (Supplementary Fig. 4). The highest number of foci was observed when the MMR-dependent G2 arrest was activated. There were no RAD51 foci in the MMR-deficient 293TLα− cells at any time point after the treatment, indicating that the damage created by MNNG was converted to recombination intermediates exclusively in MMR-proficient cells. We noted earlier (Stojic et al. 2004) that, like RAD51, RPA also forms MMR-dependent MNNG-induced foci. In order to learn whether the two polypeptides were concentrated around the same structures in the nuclei of treated cells, we stained MMR-proficient HeLa cells with antibodies against both RAD51 and RPA. (HeLa rather than 293TLα cells were used here for technical reasons.) As shown in Figure 6B, the number of foci containing these proteins peaked 48 h after MNNG treatment, and the majority colocalized. RAD51 and RPA were reported to colocalize at ssDNA regions earlier, and this phenomenon was proposed to be indicative of their common involvement in recombination repair (Raderschall et al. 1999). Given that none of the latter phenomena were detectable in the treated cells after the first S phase, but that HR is required for the cells to cross the mitotic boundary, we conclude that the events involving HR factors in the first and second cell cycles after MNNG treatment differ.
MNNG-induced cell cycle arrest can be abrogated by removal of MeG after the first S phase
Up to this point we could show that the MNNG-induced cell cycle arrest is activated by MeG (Fig. 1) and that the presence of this modified base in DNA does not retard replication (Fig. 2), but that the newly synthesized DNA contains persistent MMR-dependent gaps (Fig. 3) that may be constantly undergoing metabolism, as suggested by the association of PCNA with chromatin outside of S phase (Fig. 4). We also showed that HR is required for transition of the treated cells into the second cell cycle (Fig. 5) and that mammalian cells that undergo a cell cycle arrest after the second S phase mostly die, but that the surviving fraction contains an elevated frequency of SCEs (Fig. 6), indicative of double-strand break repair at collapsed replication forks. Taken together, this evidence is consistent with our hypothesis that unsuccessful processing of MeG/T and MeG/C mispairs by MMR leaves single-stranded gaps in the DNA after the first round of DNA replication, which can be filled neither by polymerases nor by HR, due to persisting MeG residues. Once these gaps reach replication forks in the second S phase, they give rise to double-strand breaks, which cause the replication forks to collapse. Their rescue requires HR again, which generates SCEs.
Because the above hypothesis posits that MeG residues represent the only barrier that prevents polymerases and/or HR from repairing the persistent single-stranded gaps, we removed this modified base from the DNA after the first or second S phase and followed the progression of the cells through the cell cycle. In a control experiment, we synchronized HeLa cells at the G1/S boundary by a 16-h treatment with 2 mM HU. As shown in Figure 7A, when the cells were released into HU-free medium, they re-entered the cell cycle and reverted to an asynchronous population 24–36 h later. During this time, the levels of MGMT protein remained largely constant. When the cells were released into a medium containing 0.1 μM MNNG (Fig. 7B), the MGMT levels remained unchanged and were presumably sufficient to revert the MeG residues back to Gs, as the cells continued to cycle similarly to the untreated control. However, when the cells were pretreated with BG for 16 h, the amount of MGMT was notably reduced (Fig. 7C) and the treated cells arrested in the G2/M phase of the second cell cycle, as indicated by the cell cycle profiles after 36 and 60 h.
MGMT is a small protein (∼25 kDa); correspondingly, after the removal of BG from the medium, its levels are rapidly replenished (Pieper 1997). We made use of this finding to restore MeG repair in the treated cells. We removed BG from the medium 10 h after release from the HU block, when the cells passed the first S phase, but have not yet entered the second cell cycle. As shown in Figure 7A, the period between BG removal (10 h) and the time point when the arrest normally became effective (36 h) allowed for the recovery of normal MGMT levels and, presumably, MeG repair. Importantly, the cells returned to a normal cycling profile (see 60-h time point). When BG was removed from the medium 36 h after release from HU—i.e., when the checkpoint had already been activated (Fig. 7A)—the cells remained arrested. This experiment shows that MeG is the only lesion in the DNA of MNNG-treated cells that must persist in order for the checkpoint to be activated. But the experiment also shows that MeG does not signal to the checkpoint machinery directly; the checkpoint would be activated sooner if it did. Most importantly, the data show that the lesion(s) triggering the checkpoint can be repaired rapidly and efficiently once MeG is removed from DNA. This would argue more in favor of single-stranded gaps than complex recombination intermediates.
Discussion
The cytotoxicity of SN1-type methylating agents has been unambiguously linked with the modification of guanine on the O6 position. However, how MeG brings about cell death has been subject to discussion. The so-called “signaling hypothesis” suggested that MeG-containing mispairs arising during the first DNA replication are bound by MutSα and that this is sufficient to activate cell cycle arrest (Fishel 1998; Yoshioka et al. 2006). The “processing hypothesis” posits that it is the futile processing of these mispairs by MMR that gives rise to DNA intermediates, which then signal to the checkpoint machinery (Karran and Bignami 1996). In the present study, we show that the mere presence of MeG in the genomic DNA of MMR-proficient cells does not bring about cell killing, even if the cells are allowed to undergo one round of replication, during which MeG/C and MeG/T pairs can form; to activate the checkpoint, these mispairs must be processed.
The finding that MSH3 and MSH6 carry PCNA-binding motifs and interact with this polymerase processivity factor in vitro and in vivo implied that MMR is intimately linked with the replication machinery (Clark et al. 2000; Flores-Rozas et al. 2000; Kleczkowska et al. 2001). The ability of MMR factors to halt the progress of the replicating polymerase upon mismatch detection might liberate the 3′ end of the newly synthesized strand and thus provide the excision machinery with an entry point, where degradation of the error-containing strand could commence. In this scenario, iterative attempts of the replicative polymerase to find a perfect match for MeG would be expected to stall, or at least slow down, the progression of the replication fork. DNA synthesis across MeG has indeed been shown to retard replicative polymerases in vitro (Haracska et al. 2000; York and Modrich 2006); however, neither the early experiments (Plant and Roberts 1971) nor our measurements of gross replication rates in mammalian cells (Fig. 2), nor examination of the progression of replication forks from a defined origin in yeast cells (Supplementary Fig. 1), detected a MMR-dependent reduction of replication rates in the first S phase after MNNG treatment. This suggests that MMR acting on MeG is a post-replicative process, as shown recently for translesion synthesis (Lopes et al. 2006).
Futile attempts of DNA polymerase(s) to incorporate the correct nucleotide opposite MeG were proposed to result in the generation of ssDNA gaps, the formation of which has recently been inferred in biochemical experiments (York and Modrich 2006). Our EM analysis of RIs isolated from MNNG-treated MMR-proficient yeast and mammalian cells shows that such ssDNA gaps exist also in vivo. That the gaps were often far from the fork and persisted in the replicated DNA for a long time (Fig. 3D,E) indicated that they did not block fork progression, but that they could not be effectively repaired. This might be expected, given that every attempt of a polymerase to fill the gap will be aborted by MMR, because repair synthesis will regenerate MeG/T or MeG/C mispairs. The findings that the internal gaps are far from the replication forks further strengthens the hypothesis that iterative attempts to fill these gaps are carried out behind the replication forks. It is possible that the dispersed staining of chromatin-bound PCNA observed in MNNG-treated cells, which contrasted with the clear foci (Dimitrova and Gilbert 2000) observed during an unperturbed S phase (Fig. 4), represents these iterative cycles of DNA excision and resynthesis at MeG sites.
How might ssDNA gaps persist through two cell cycles? Previous reports showed that HR was activated upon MNNG treatment of both mammalian and yeast cells in an MMR-dependent manner (Zhang et al. 2000; Cejka et al. 2005). Our experiments demonstrate that the recombination events are not merely the consequence of strand break formation linked with MNNG treatment, but that they are required for survival and progression across the mitotic boundary, as documented by the sensitivity of HR-deficient mammalian cells to MNNG-induced killing (Fig. 5A) and by the failure of HR-deficient yeast and mammalian cells to progress to the second cell cycle after MNNG treatment (Fig. 5B,D; see also Tsaryk et al. 2006). Although HR-proficient yeast cells were substantially more resistant to MNNG treatment than mammalian cells, and although they did not arrest at G2/M two cell cycles after treatment, it became evident from the cell cycle analysis that their G2 phase was significantly longer than that of untreated cells even three cell cycles post-treatment. Such prolonged G2/M phase may be a manifestation of ongoing repair, which, coupled with the efficient HR in Saccharomyces cerevisiae, may explain the relative resistance of yeast cells to killing by MNNG.
The MMR-dependent formation of ssDNA gaps after MNNG treatment is a common feature in both yeast and mammalian cells. In unmodified DNA, such single-stranded gaps would be repaired by unscheduled DNA synthesis or recombination (Scully et al. 2000). However, in MNNG-treated cells, these events would be reversed or aborted by MMR, given that both the latter processes, similarly to repair synthesis during MMR, will give rise to MeG/C and MeG/T mispairs. We therefore postulated that the MeG-containing gaps might be bound by RPA and/or by RAD51 or its paralogs, which are known to bind both ssDNA nicks and gaps in mammalian cells (Masson et al. 2001). This might protect them from further processing and thus enable them to progress to the subsequent cell division. The absence of RAD51 and RPA foci in the first cell cycle post-treatment (Fig. 6B) might be explained by the fact that the gaps are generally shorter than 200 nt (Fig. 3) and thus that the number of RAD51 and/or RPA molecules bound to them might be too low to appear as foci (Raderschall et al. 1999).
The small size of the detected ssDNA gaps may also explain why they fail to trigger a cell cycle arrest already after the first S phase; it is likely that the amount of exposed ssDNA lies below the threshold that has to be exceeded before a DNA replication checkpoint is activated (Shimada et al. 2002). In our earlier work, we detected transient DNA damage signaling (Stojic et al. 2004), and the present study detected a delay (Fig. 2B,F) in the first cycle after MNNG treatment. These events might be triggered by the direct detection of mismatches by MutSα (Yoshioka et al. 2006), the binding of RPA to unrepaired ssDNA gaps, or the formation of more complex intermediates primed by these gaps, which might interfere with proper DNA segregation. However, the signal generated by these events is either too low to activate the checkpoint proper, or is attenuated, possibly through the binding of recombination factors to ssDNA (Fig. 8), which has been shown to prevent checkpoint activation in the case of irreparable resected DSBs (Lee et al. 2003). Only during the second replication would these primary lesions (ssDNA gaps and/or structures derived from them) give rise to full checkpoint activation, cell cycle arrest, and cell death.
Figure 8.
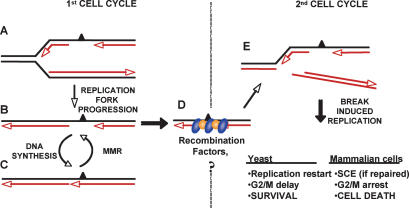
Model for the roles of replication, MMR, and recombination in DNA transactions induced by MeG. (A) When replication forks encounter MeG in the template DNA of MMR-proficient cells, they insert C or T and proceed with replication. The MeG-containing mispair activates MMR, which degrades the newly synthesized strand up to and some distance past the MeG residue. (B,C) This gap cannot be filled in, because MMR repeatedly inhibits post-replicative gap repair. In the presence of functional recombination, the gaps are protected and progress to the next cell cycle (D), where they cause replication fork collapse (E). The collapsed replication forks can be restored with the help of HR, which leads to cell survival, at a cost of higher SCE levels. Cells that fail to rescue the forks arrest in G2/M and subsequently die because of their inability to restart replication.
We cannot formally demonstrate that these gaps persist through mitosis. However, their persistence until the second round of replication is supported by three lines of evidence. First, gaps in template DNA should bring about replication fork collapse, which is consistent with our earlier findings showing that MNNG induces an ATR-dependent G2 arrest after the second S phase (Stojic et al. 2004). Second, when replication forks collapse as a result of a double-strand break, the ends of the chromatid are resected to give rise to long ssDNA filaments, which are bound by many RAD51 and/or RPA molecules. This agrees with the appearance of large foci of these two proteins after the second S phase (Fig. 6B). Third, collapsed replication forks give rise to two linear DNA molecules (Fig. 8E), which would not be isolated as RIs by our experimental procedure. Indeed, analysis of RIs isolated from the second cell cycle of MNNG-treated human cells failed to reveal the presence of abnormal DNA structures (data not shown). Taken together, this evidence lends support to the hypothesis that the MMR and MNNG dose-dependent reduction of DNA synthesis rates in the second cell cycle (Fig. 2) is indeed linked to replication fork collapse caused by persistent gaps in template DNA.
In wild-type yeast cells, the high efficiency of HR overcomes the cytotoxic effects of MNNG treatment, and the only consequence of the damage is a delay in G2/M. In contrast, mammalian cells cannot cope with the high number of collapsed replication forks (Figs. 2, 8), and the small proportion of cells that recover from the G2/M arrest in the second cell cycle display elevated levels of SCEs (Kaina et al. 1997), which are strictly dependent on MMR (Fig. 6B). While SCEs are considered to arise through HR-mediated processing of stalled or collapsed replication forks (Richardson et al. 1998; Johnson and Jasin 2000), recombination-dependent processing of ssDNA gaps, proposed to result in the formation of double Holliday junctions, does not give rise to SCEs (Helleday 2003). Given that SCEs were observed only after the second S phase in the 293T La+ cells provides further support for the hypothesis that MNNG treatment of MGMT-deficient cells leads to the MMR-dependent generation of persistent single-stranded gaps in newly replicated DNA.
In cells expressing low amounts of MGMT, such as tumors in which the gene is epigenetically silenced (Jacinto and Esteller 2007), the cytotoxicity of SN1-type methylating agents was linked with the persistence of MeG until DNA replication, where unsuccessful attempts of the MMR system to correct MeG/C and MeG/T mispairs lead to cell death (Karran 2001). In our previous study, we postulated that unsuccessful MMR leaves irreparable ssDNA gaps in the DNA after the first S phase, which give rise to double-strand breaks during the subsequent replication (Stojic et al. 2004). In the present study, we substantiate this hypothesis experimentally by demonstrating the existence of these gaps. Moreover, we show that HR permits the cells with incompletely repaired DNA to enter the subsequent cell cycle, where these discontinuities cause replication fork collapse and cell cycle arrest. Although most mammalian cells never recover from this block, some are rescued by HR-mediated repair of the collapsed replication forks, which gives rise to SCEs. Finally, the ready abrogation of the cell cycle arrest brought about by re-expression of MGMT after the first S phase argues against the existence of complex irreparable recombination intermediates arising after DNA replication; rather, it shows that MeG represents the only hurdle to the repair or resolution of these structures. In an attempt to provide experimental support for this hypothesis, we allowed the MNNG-treated cells to re-express MGMT before the second S phase and thus remove these modifications. As shown in Figure 7D, this abolished the G2/M arrest, and the cells continued to cycle. The most likely explanation for this finding is that removal of MeG from the gaps or recombination intermediates permits their immediate repair.
SN1-type methylating agents are in widespread use in cancer chemotherapy. By elucidating their mode of action, and in particular by demonstrating the requirement for HR in the processing of DNA damage induced by these agents, our work implies that inhibition of HR would substantially increase the efficacy of this important class of chemotherapeutics.
Materials and methods
Cell lines and chemicals
The 293T Lα cell line was established in our laboratory and was propagated as described (Cejka et al. 2003). The HeLa and HeLaMR (a kind gift of Margherita Bignami) cells were maintained in DMEM (OmniLab) supplemented with 10% fetal calf serum (FCS; Life Technologies), penicillin (100 U/mL), and streptomycin (100 μg/mL). The HR-deficient Chinese hamster cell line IRS1 was mutated in XRCC2; the defect was corrected in the IRS1/XRCC2 line by a stable transfection with human XRCC2 cDNA. Both lines were kindly provided by John Thacker. The XRCC3-deficient cells and their HR-proficient parental cell line AA8 were kindly provided by Orlando Schärer. All four lines were maintained in DMEM:F10 (1:1; OmniLab), supplemented with 10% FCS. To inhibit MGMT activity, the cells were pretreated with 10 μM BG. MNNG, BG, HU, and 6TG were purchased from Sigma, and Dox was purchased from Clontech.
The yeast strains used in this study were described previously (Cejka et al. 2005). They were grown in YPD at 25°C.
Cell synchronization and cell cycle analysis
The yeast cells were synchronized in G1 phase by adding 2 μg/mL α-factor and, after centrifugation, were released into a medium with or without 3 μM MNNG.
The mammalian cells were synchronized with 2 mM HU for 16 h. They were washed twice with PBS and incubated in fresh medium with or without MNNG and BG. At the indicated time points, cells were trypsinized and processed for cell cycle analyses as described previously (Stojic et al. 2004). Cell cycle analyses were performed using a Beckman Coulter FC 500 cytometer.
Antibodies and immunoblotting
Immunoblotting and total protein extractions were performed as described previously (Cejka et al. 2003). The anti-β-tubulin (D-10) antibody was from Santa Cruz Biotechnology, and the anti-MGMT antibody (Clone MT 23.2) was from GeneTex.
Replication rates
The cells were plated and allowed to attach for 24 h. [14C] Thymidine [20 nCi/mL] was added, and the cells were allowed to grow for an additional 24 h. After washing with PBS, the cells were incubated in growth medium for 10 h, and 2 mM HU was added for an additional 16 h. At this time point (Release), the cells were washed with PBS and incubated with normal growth medium without (Control) or with MNNG or cisplatin. Fifteen minutes before the indicated time points, 2.5 μCi/mL [3H] thymidine was added. One-fifth of each sample was processed for cell cycle analysis, and the rest was processed for scintillation counting. The cells were collected by centrifugation, the pellets were resuspended in 280 μL of ice-cold PBS, and 720 μL of ice-cold methanol were added dropwise. The cells were pelleted and resuspended in 200 μL of PBS, and 5 mL of scintillation liquid (Optiphase “Hisafe” 2; PerkinElmer) were added. The samples were incubated overnight at +4°C before scintillation counting. DNA synthesis was estimated as the ratio of [3H]/[14C].
EM analysis of total genomic DNA
In vivo psoralen cross-linking, isolation of total genomic DNA, and enrichment of the RIs from yeast cells were performed as described previously (Lopes et al. 2006). For EM analysis of the mammalian RIs, 293T Lα cells were seeded and synchronized with 2 mM HU as described above. The cells were released into fresh medium without (Not Treated, NT) or with 5 μM MNNG and were collected by trypsinization after 4 h. At this time point, the majority of the cells were in mid-S phase, as assessed by cell cycle analysis (Fig. 2; data not shown). Approximately 107 cells were resuspended in 10 mL of ice-cold PBS and transferred to a 6-cm dish to which 10 μg/mL trimethylpsoralen (TMP) were added for 5 min in the dark. The cells were irradiated with 366-nm monochromatic light for 5 min, at a distance of ∼1 cm from the lamps (Stratalinker; Stratagene). The TMP addition in the dark and irradiation were repeated three more times. Subsequently, the cells were spun down, and genomic DNA was isolated using the QIAamp DNA blood minikit (Qiagen), according to the manufacturer’s instructions. The DNA was digested with PvuII, and RIs were enriched using the same procedure as for the yeast DNA.
Immunofluorescence staining
For PCNA staining with permabilization before fixation, cells grown on glass coverslips were synchronized with double-thymidine block as described (Stojic et al. 2004), treated or mock-treated with MNNG, and incubated for the indicated times. The slides were immersed for 5 min. in ice-cold permeabilization buffer (10 mM Tris at pH 7.6, 2.5 mM MgCl2, 0.5% NP-40, 0.2 mM PMSF) before fixation in ice-cold methanol. This treatment removes all soluble proteins from the nucleoplasm, while the chromatin-bound fraction is retained. The slides were subsequently stained with an anti-PCNA antibody (PC-10; Santa Cruz Biotechnology). Cells stained with RAD51 (rabbit antibody; Pharmingen) and RPA (mouse anti-p32; Oncogene) antibodies were fixed in 3.7% formaldehyde and permeabilized with 0.2% Triton-X, and the incubation with antibodies was carried out as for PCNA. After washing, the cells were incubated at 37°C with TR-conjugated anti-mouse antibody (Abcam) and anti-rabbit FITC (Sigma). Cell nuclei were counterstained with DAPI and mounted in anti-fade (Molecular Probes). Images were acquired on Olympus IX81 microscope, equipped with appropriate filter sets and using CellR imaging software.
SCE assay
To differentially label sister chromatids, the cells were incubated for 48 h with 10 μM BrdU (two cell cycles). Assessing the effect of MNNG was accomplished by the addition of 0.2 μM MNNG 24 or 48 h before harvesting. Preparation of the mitotic spreads and subsequent staining were carried out as described (Kaina and Aurich 1985). The number of SCE events shown in Figure 6A was counted in 50 metaphases.
Clonogenic survival assay
The cells were treated with the indicated concentrations of MNNG for 2 h, trypsinized, and counted. Serial dilutions of control and treated cells were plated and left for 14–20 d, after which the colonies were stained with 10% Giemsa and counted.
Acknowledgments
We thank Lovorka Stojic and Petr Cejka for helpful discussions, and Margherita Bignami, Orlando Schärer, and John Thacker for the kind gift of the cell lines. We are grateful to the Electron Microscopy Center of the ETH Zürich for technical support. We also gratefully acknowledge the financial support of the Bonizzi-Theler Stiftung, the European Community, and the Swiss National Science Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.455407
References
- Berardini M., Mazurek A., Fishel R., Mazurek A., Fishel R., Fishel R. The effect of O6-methylguanine DNA adducts on the adenosine nucleotide switch functions of hMSH2–hMSH6 and hMSH2–hMSH3. J. Biol. Chem. 2000;275:27851–27857. doi: 10.1074/jbc.M003589200. [DOI] [PubMed] [Google Scholar]
- Cejka P., Stojic L., Mojas N., Russell A.M., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J., Stojic L., Mojas N., Russell A.M., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J., Mojas N., Russell A.M., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J., Russell A.M., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J., Cannavo E., di Pietro M., Marra G., Jiricny J., di Pietro M., Marra G., Jiricny J., Marra G., Jiricny J., Jiricny J. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Mojas N., Gillet L., Schar P., Jiricny J., Mojas N., Gillet L., Schar P., Jiricny J., Gillet L., Schar P., Jiricny J., Schar P., Jiricny J., Jiricny J. Homologous recombination rescues mismatch-repair-dependent cytotoxicity of S(N)1-type methylating agents in S. cerevisiae. Curr. Biol. 2005;15:1395–1400. doi: 10.1016/j.cub.2005.07.032. [DOI] [PubMed] [Google Scholar]
- Clark A.B., Valle F., Drotschmann K., Gary R.K., Kunkel T.A., Valle F., Drotschmann K., Gary R.K., Kunkel T.A., Drotschmann K., Gary R.K., Kunkel T.A., Gary R.K., Kunkel T.A., Kunkel T.A. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Gilbert D.M., Gilbert D.M. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2000;2:686–694. doi: 10.1038/35036309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M.E., Roy S.K., Garbiras B.J., Helft P., Paras P., Chae M.-Y., Moschel R.C., Pegg A.E., Roy S.K., Garbiras B.J., Helft P., Paras P., Chae M.-Y., Moschel R.C., Pegg A.E., Garbiras B.J., Helft P., Paras P., Chae M.-Y., Moschel R.C., Pegg A.E., Helft P., Paras P., Chae M.-Y., Moschel R.C., Pegg A.E., Paras P., Chae M.-Y., Moschel R.C., Pegg A.E., Chae M.-Y., Moschel R.C., Pegg A.E., Moschel R.C., Pegg A.E., Pegg A.E. O6-Alkylguanine-DNA alkyltransferase inactivation by ester prodrugs of O6-benzylguanine derivatives and their rate of hydrolysis by cellular esterases. Biochem. Pharmacol. 1998;55:1701–1709. doi: 10.1016/s0006-2952(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Duckett D.R., Drummond J.T., Murchie A.I.H., Reardon J.T., Sancar A., Lilley D.M.J., Modrich P., Drummond J.T., Murchie A.I.H., Reardon J.T., Sancar A., Lilley D.M.J., Modrich P., Murchie A.I.H., Reardon J.T., Sancar A., Lilley D.M.J., Modrich P., Reardon J.T., Sancar A., Lilley D.M.J., Modrich P., Sancar A., Lilley D.M.J., Modrich P., Lilley D.M.J., Modrich P., Modrich P. Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes & Dev. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H., Clark D., Kolodner R.D., Clark D., Kolodner R.D., Kolodner R.D. Proliferating cell nuclear antigen and Msh2p–Msh6p interact to form an active mispair recognition complex. Nat. Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- Haracska L., Prakash S., Prakash L., Prakash S., Prakash L., Prakash L. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Henry-Mowatt J., Jackson D., Masson J.Y., Johnson P.A., Clements P.M., Benson F.E., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Jackson D., Masson J.Y., Johnson P.A., Clements P.M., Benson F.E., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Masson J.Y., Johnson P.A., Clements P.M., Benson F.E., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Johnson P.A., Clements P.M., Benson F.E., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Clements P.M., Benson F.E., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Benson F.E., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Thompson L.H., Takeda S., West S.C., Caldecott K.W., Takeda S., West S.C., Caldecott K.W., West S.C., Caldecott K.W., Caldecott K.W. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell. 2003;11:1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- Jacinto F.V., Esteller M., Esteller M. MGMT hypermethylation: A prognostic foe, a predictive friend. DNA Repair (Amst.) 2007;6:1155–1160. doi: 10.1016/j.dnarep.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Johnson R.D., Jasin M., Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N.J., Cox R., Thacker J., Cox R., Thacker J., Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res./DNA Repair Reports. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Kaina B. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: A long road traveled and still a far way to go. Cytogenet. Genome Res. 2004;104:77–86. doi: 10.1159/000077469. [DOI] [PubMed] [Google Scholar]
- Kaina B., Aurich O., Aurich O. Dependency of the yield of sister-chromatid exchanges induced by alkylating agents on fixation time: Possible involvement of secondary lesions in sister-chromatid exchange induction. Mutat. Res. 1985;149:451–461. doi: 10.1016/0027-5107(85)90163-0. [DOI] [PubMed] [Google Scholar]
- Kaina B., Ziouta A., Ochs K., Coquerelle T., Ziouta A., Ochs K., Coquerelle T., Ochs K., Coquerelle T., Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex−, Mex+ and methylation-tolerant mismatch repair compromised cells: Facts and models. Mutat. Res. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- Karran P. Thiopurines, DNA damage, DNA repair and therapy-related cancer. Br. Med. Bull. 2006;79-80:153–170. doi: 10.1093/bmb/ldl020. [DOI] [PubMed] [Google Scholar]
- Karran P., Bignami M., Bignami M. Drug-related killings: A case of mistaken identity. Chem. Biol. 1996;3:875–879. doi: 10.1016/s1074-5521(96)90175-1. [DOI] [PubMed] [Google Scholar]
- Kleczkowska H.E., Marra G., Lettieri T., Jiricny J., Marra G., Lettieri T., Jiricny J., Lettieri T., Jiricny J., Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes & Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Pellicioli A., Vaze M.B., Sugawara N., Malkova A., Foiani M., Haber J.E., Pellicioli A., Vaze M.B., Sugawara N., Malkova A., Foiani M., Haber J.E., Vaze M.B., Sugawara N., Malkova A., Foiani M., Haber J.E., Sugawara N., Malkova A., Foiani M., Haber J.E., Malkova A., Foiani M., Haber J.E., Foiani M., Haber J.E., Haber J.E. Yeast Rad52 and Rad51 recombination proteins define a second pathway of DNA damage assessment in response to a single double-strand break. Mol. Cell. Biol. 2003;23:8913–8923. doi: 10.1128/MCB.23.23.8913-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J.M., Foiani M., Sogo J.M., Sogo J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Masson J.Y., Tarsounas M.C., Stasiak A.Z., Stasiak A., Shah R., McIlwraith M.J., Benson F.E., West S.C., Tarsounas M.C., Stasiak A.Z., Stasiak A., Shah R., McIlwraith M.J., Benson F.E., West S.C., Stasiak A.Z., Stasiak A., Shah R., McIlwraith M.J., Benson F.E., West S.C., Stasiak A., Shah R., McIlwraith M.J., Benson F.E., West S.C., Shah R., McIlwraith M.J., Benson F.E., West S.C., McIlwraith M.J., Benson F.E., West S.C., Benson F.E., West S.C., West S.C. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes & Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva O.K., Petrini J.H.J., Petrini J.H.J. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowosielska A., Smith S.A., Engelward B.P., Marinus M.G., Smith S.A., Engelward B.P., Marinus M.G., Engelward B.P., Marinus M.G., Marinus M.G. Homologous recombination prevents methylation-induced toxicity in Escherichia coli. Nucleic Acids Res. 2006;34:2258–2268. doi: 10.1093/nar/gkl222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan P., Wilson C., Townsend S., Thacker J., Wilson C., Townsend S., Thacker J., Townsend S., Thacker J., Thacker J. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem. 2001;276:22148–22153. doi: 10.1074/jbc.M102396200. [DOI] [PubMed] [Google Scholar]
- Pieper R.O. Understanding and manipulating O6-methylguanine-DNA methyltransferase expression. Pharmacol. Ther. 1997;74:285–297. doi: 10.1016/s0163-7258(97)00003-x. [DOI] [PubMed] [Google Scholar]
- Plant J.E., Roberts J.J., Roberts J.J. A novel mechanism for the inhibition of DNA synthesis following methylation: The effect of N-methyl-N-nitrosourea on HeLa cells. Chem. Biol. Interact. 1971;3:337–342. doi: 10.1016/0009-2797(71)90013-5. [DOI] [PubMed] [Google Scholar]
- Raderschall E., Golub E.I., Haaf T., Golub E.I., Haaf T., Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C., Moynahan M.E., Jasin M., Moynahan M.E., Jasin M., Jasin M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Puget N., Vlasakova K., Puget N., Vlasakova K., Vlasakova K. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene. 2000;19:6176–6183. doi: 10.1038/sj.onc.1203971. [DOI] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S.M., Pasero P., Gasser S.M., Gasser S.M. ORC and the intra-S-phase checkpoint: A threshold regulates Rad53p activation in S phase. Genes & Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L., Mojas N., Cejka P., Di Pietro M., Ferrari S., Marra G., Jiricny J., Mojas N., Cejka P., Di Pietro M., Ferrari S., Marra G., Jiricny J., Cejka P., Di Pietro M., Ferrari S., Marra G., Jiricny J., Di Pietro M., Ferrari S., Marra G., Jiricny J., Ferrari S., Marra G., Jiricny J., Marra G., Jiricny J., Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes & Dev. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaryk R., Fabian K., Thacker J., Kaina B., Fabian K., Thacker J., Kaina B., Thacker J., Kaina B., Kaina B. Xrcc2 deficiency sensitizes cells to apoptosis by MNNG and the alkylating anticancer drugs temozolomide, fotemustine and mafosfamide. Cancer Lett. 2006;239:305–313. doi: 10.1016/j.canlet.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Wyatt M.D., Pittman D.L., Pittman D.L. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Scherer S.J., Shell S.S., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W., Scherer S.J., Shell S.S., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W., Shell S.S., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W., Kucherlapati R., Kolodner R.D., Edelmann W., Kolodner R.D., Edelmann W., Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- York S.J., Modrich P., Modrich P. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. J. Biol. Chem. 2006;281:22674–22683. doi: 10.1074/jbc.M603667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K.-I., Yoshioka Y., Hsieh P., Yoshioka Y., Hsieh P., Hsieh P. ATR kinase activation mediated by MutSα and MutLα in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Marra G., Jiricny J., Maher V.M., McCormick J.J., Marra G., Jiricny J., Maher V.M., McCormick J.J., Jiricny J., Maher V.M., McCormick J.J., Maher V.M., McCormick J.J., McCormick J.J. Mismatch repair is required for O6-methylguanine-induced homologous recombination in human fibroblasts. Carcinogenesis. 2000;21:1639–1646. doi: 10.1093/carcin/21.9.1639. [DOI] [PubMed] [Google Scholar]
- Zhukovskaya N., Branch P., Aquilina G., Karran P., Branch P., Aquilina G., Karran P., Aquilina G., Karran P., Karran P. DNA replication arrest and tolerance to DNA methylation damage. Carcinogenesis. 1994;15:2189–2194. doi: 10.1093/carcin/15.10.2189. [DOI] [PubMed] [Google Scholar]