Abstract
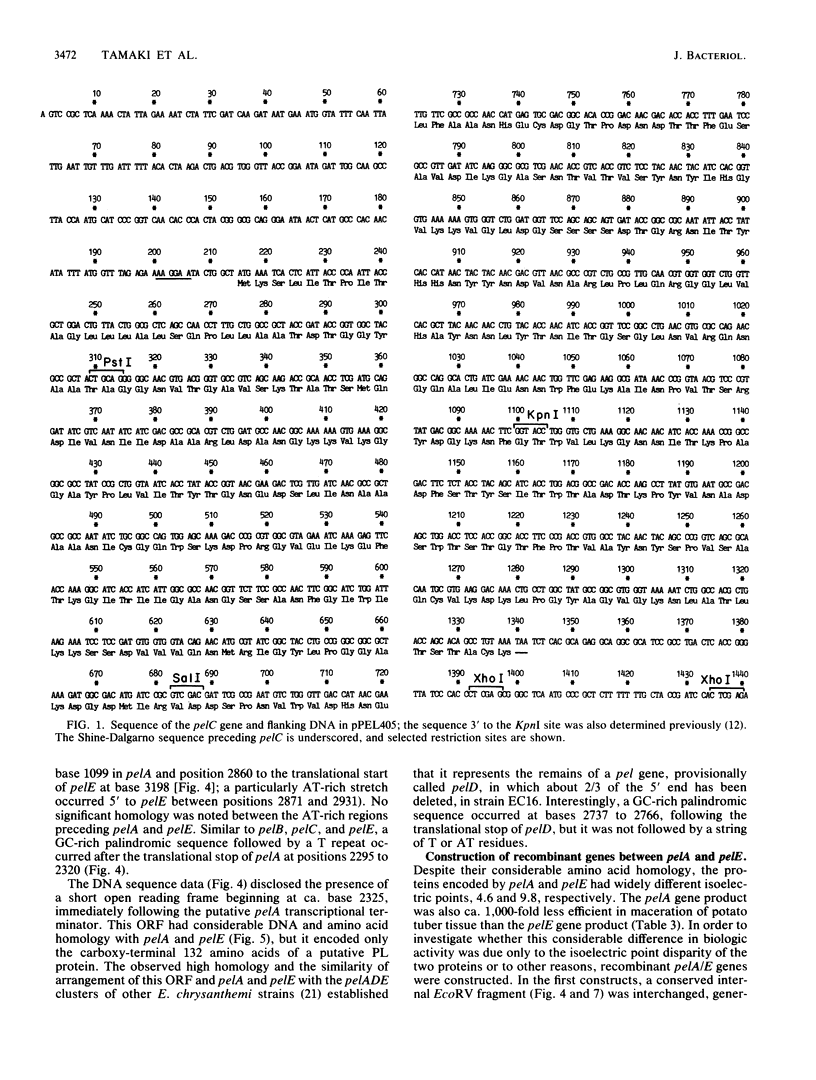
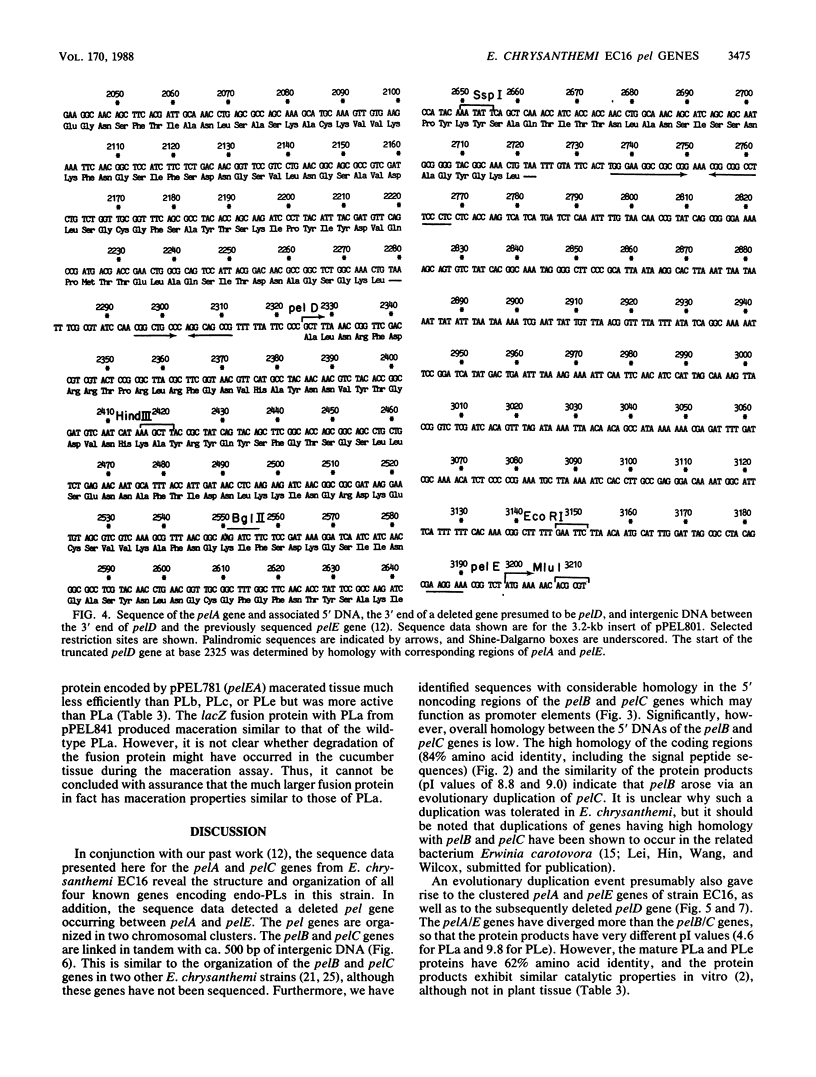
The pelA and pelC genes from Erwinia chrysanthemi EC16 were sequenced and overexpressed in Escherichia coli cells. These genes and two others from the same strain that were characterized previously encode catalytically related pectate lyase proteins that are involved with the maceration and soft-rotting of plant tissue. The pel genes of strain EC16 were organized as two loosely linked clusters, with two structurally homologous genes in each. The pelA/E cluster also contained the remains of an additional pel gene, the 5' portion of which had been removed by a prior deletion event. Each of the four functional pel genes but not the deleted one contained an efficient rho-independent transcriptional terminator after the translational stop. These and other data indicate that the pel genes are all independently regulated despite their structural homology and tandem clustered organization. Two of the genes, pelA and pelE, encoded proteins that differed greatly in their isoelectric points and ability to macerate plant tissue. A recombinant gene constructed with the 5' portion of pelE and the 3' portion of pelA yielded a chimeric protein with high pectate lyase activity but relatively low maceration activity. This result raised the possibility that the poor maceration ability of the pelA gene product may involve other properties in addition to its low isoelectric point.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barras F., Thurn K. K., Chatterjee A. K. Resolution of four pectate lyase structural genes of Erwinia chrysanthemi (EC16) and characterization of the enzymes produced in Escherichia coli. Mol Gen Genet. 1987 Sep;209(2):319–325. doi: 10.1007/BF00329660. [DOI] [PubMed] [Google Scholar]
- Barras F, Chatterjee A K. Genetic analysis of the pelA-pelE cluster encoding the acidic and basic pectate lyases in Erwinia chrysanthemi EC16. Mol Gen Genet. 1987 Oct;209(3):615–617. doi: 10.1007/BF00331172. [DOI] [PubMed] [Google Scholar]
- Chatterjee A. K., Buchanan G. E., Behrens M. K., Starr M. P. Synthesis and excretion of polygalacturonic acid trans-eliminase in Erwinia, Yersinia, and Klebsiella species. Can J Microbiol. 1979 Jan;25(1):94–102. doi: 10.1139/m79-014. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Hasan N., Szybalski W. Control of cloned gene expression by promoter inversion in vivo: construction of improved vectors with a multiple cloning site and the Ptac promoter. Gene. 1987;56(1):145–151. doi: 10.1016/0378-1119(87)90167-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Heusterspreute M., Ha Thi V., Emery S., Tournis-Gamble S., Kennedy N., Davison J. Vectors with restriction site banks. IV. pJRD184, a 3793-bp plasmid vector with 49 unique restriction sites. Gene. 1985;39(2-3):299–304. doi: 10.1016/0378-1119(85)90327-0. [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Keen N. T., Dahlbeck D., Staskawicz B., Belser W. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J Bacteriol. 1984 Sep;159(3):825–831. doi: 10.1128/jb.159.3.825-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S. Structure of two pectate lyase genes from Erwinia chrysanthemi EC16 and their high-level expression in Escherichia coli. J Bacteriol. 1986 Nov;168(2):595–606. doi: 10.1128/jb.168.2.595-606.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoujansky A., Diolez A., Boccara M., Bertheau Y., Andro T., Coleno A. Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 1985 Mar;4(3):781–785. doi: 10.1002/j.1460-2075.1985.tb03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S. P., Lin H. C., Wang S. S., Callaway J., Wilcox G. Characterization of the Erwinia carotovora pelB gene and its product pectate lyase. J Bacteriol. 1987 Sep;169(9):4379–4383. doi: 10.1128/jb.169.9.4379-4383.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manulis S., Kobayashi D. Y., Keen N. T. Molecular cloning and sequencing of a pectate lyase gene from Yersinia pseudotuberculosis. J Bacteriol. 1988 Apr;170(4):1825–1830. doi: 10.1128/jb.170.4.1825-1830.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne J. H., Schoedel C., Keen N. T., Collmer A. Multiplication and Virulence in Plant Tissues of Escherichia coli Clones Producing Pectate Lyase Isozymes PLb and PLe at High Levels and of an Erwinia chrysanthemi Mutant Deficient in PLe. Appl Environ Microbiol. 1987 Oct;53(10):2315–2320. doi: 10.1128/aem.53.10.2315-2320.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S., Van Gijsegem F., Rouve M., Kotoujansky A., Robert-Baudouy J. Organization of a pectate lyase gene family in Erwinia chrysanthemi. Gene. 1986;49(2):215–224. doi: 10.1016/0378-1119(86)90282-9. [DOI] [PubMed] [Google Scholar]
- Ried J. L., Collmer A. Comparison of pectic enzymes produced by Erwinia chrysanthemi, Erwinia carotovora subsp. carotovora, and Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol. 1986 Aug;52(2):305–310. doi: 10.1128/aem.52.2.305-310.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlemmer A. F., Ware C. F., Keen N. T. Purification and characterization of a pectin lyase produced by Pseudomonas fluorescens W51. J Bacteriol. 1987 Oct;169(10):4493–4498. doi: 10.1128/jb.169.10.4493-4498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoedel C., Collmer A. Evidence of homology between the pectate lyase-encoding pelB and pelC genes in Erwinia chrysanthemi. J Bacteriol. 1986 Jul;167(1):117–123. doi: 10.1128/jb.167.1.117-123.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wright J. J., Hayward R. S. Transcriptional termination at a fully rho-independent site in Escherichia coli is prevented by uninterrupted translation of the nascent RNA. EMBO J. 1987 Apr;6(4):1115–1119. doi: 10.1002/j.1460-2075.1987.tb04866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



