Abstract
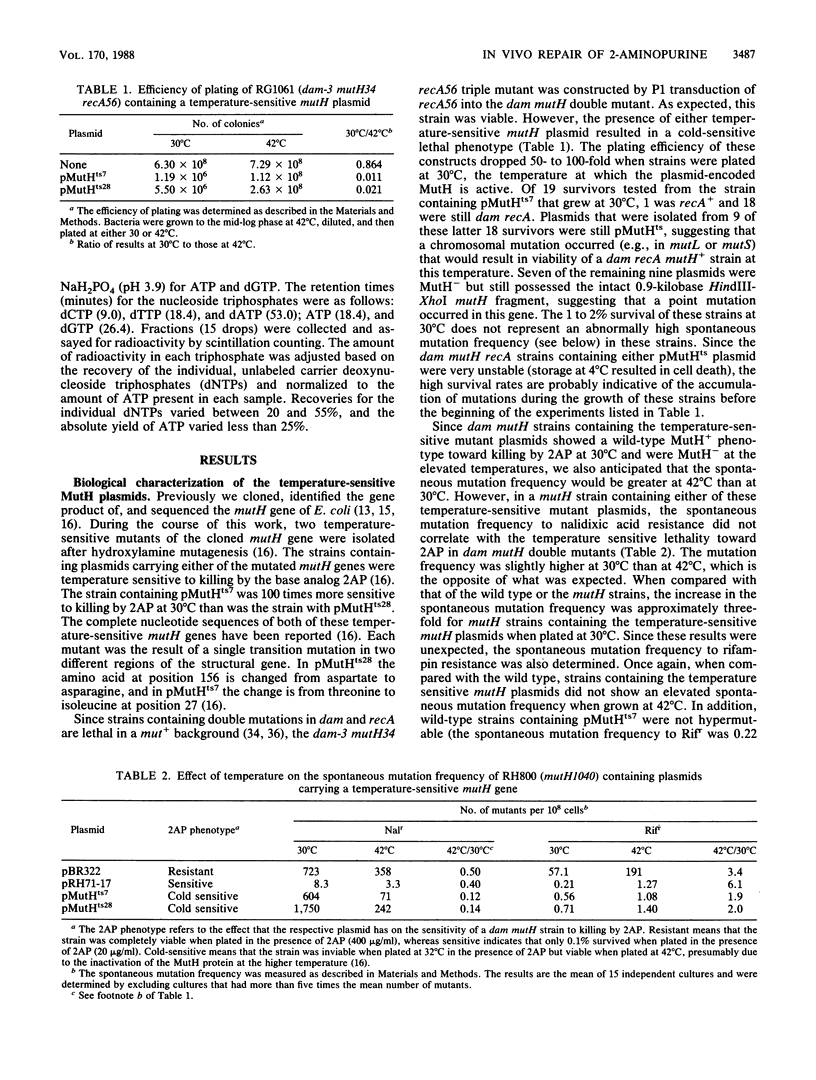
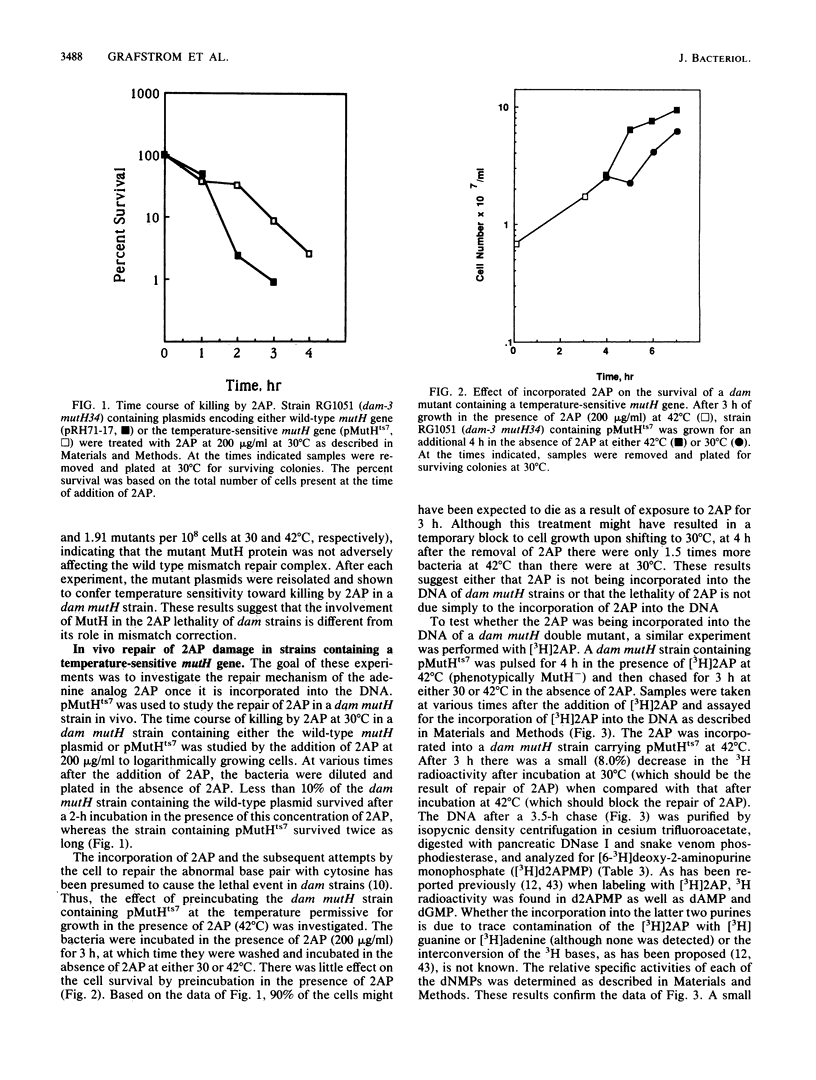
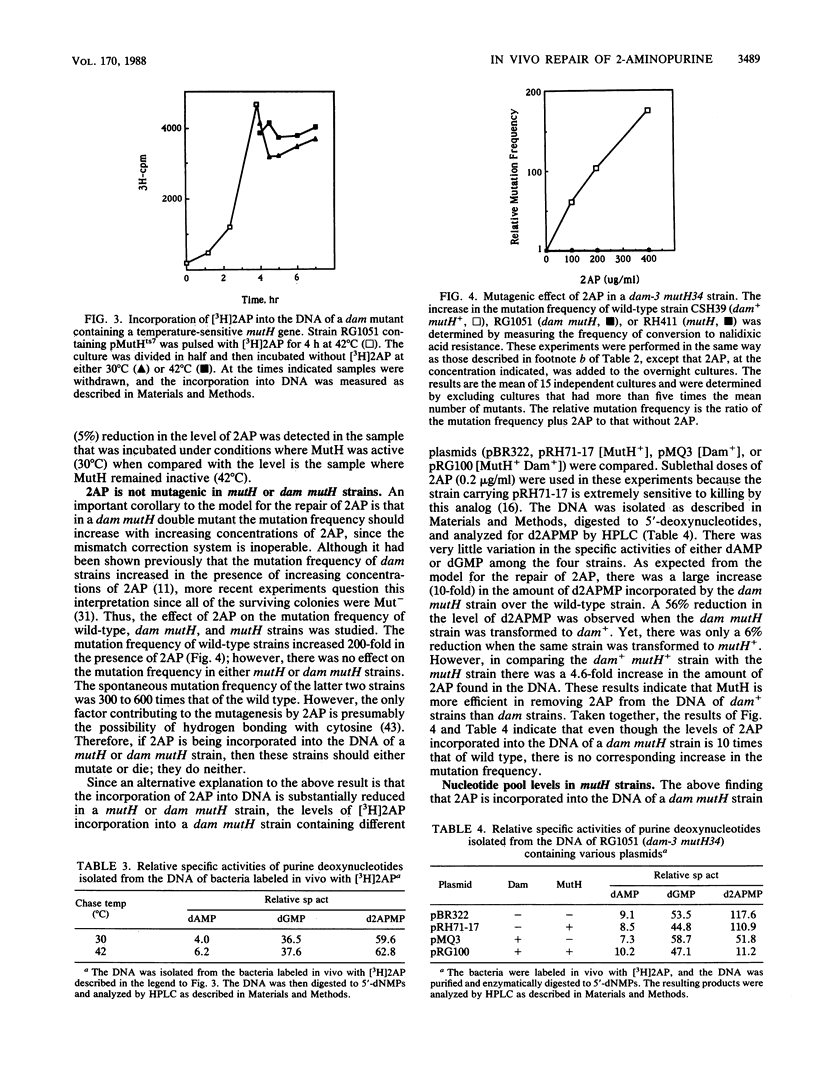
The repair of the base analog 2-aminopurine has been studied in vivo by using a temperature-sensitive mutant of the cloned mutH gene of Escherichia coli. Our results suggest that the lethal event in killing of dam mutants by 2-aminopurine does not result simply from incorporation of 2-aminopurine into the DNA and its subsequent repair. Furthermore, a 10-fold increase in the level of 2-aminopurine incorporated into the DNA of a dam mutH double mutant has little effect on the mutation frequency of this strain. An alternative mechanism for the mutagenicity of 2-aminopurine in E. coli is proposed.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arraj J. A., Marinus M. G. Phenotypic reversal in dam mutants of Escherichia coli K-12 by a recombinant plasmid containing the dam+ gene. J Bacteriol. 1983 Jan;153(1):562–565. doi: 10.1128/jb.153.1.562-565.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Ames B. N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982 Aug 25;257(16):9759–9769. [PubMed] [Google Scholar]
- Caras I. W., MacInnes M. A., Persing D. H., Coffino P., Martin D. W., Jr Mechanism of 2-aminopurine mutagenesis in mouse T-lymphosarcoma cells. Mol Cell Biol. 1982 Sep;2(9):1096–1103. doi: 10.1128/mcb.2.9.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton L. K., Goodman M. F., Branscomb E. W., Galas D. J. Error induction and correction by mutant and wild type T4 DNA polymerases. Kinetic error discrimination mechanisms. J Biol Chem. 1979 Mar 25;254(6):1902–1912. [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., Siegel E. C., Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986 Mar 20;188(2):147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Goodman M. F., Hopkins R., Gore W. C. 2-Aminopurine-induced mutagenesis in T4 bacteriophage: a model relating mutation frequency to 2-aminopurine incorporation in DNA. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4806–4810. doi: 10.1073/pnas.74.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Hoess R. H. Cloning of mutH and identification of the gene product. Gene. 1983 May-Jun;22(2-3):245–253. doi: 10.1016/0378-1119(83)90109-9. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Hoess R. H. Nucleotide sequence of the Escherichia coli mutH gene. Nucleic Acids Res. 1987 Apr 10;15(7):3073–3084. doi: 10.1093/nar/15.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Yuan R., Hamilton D. L. The characteristics of DNA methylation in an in vitro DNA synthesizing system from mouse fibroblasts. Nucleic Acids Res. 1985 Apr 25;13(8):2827–2842. doi: 10.1093/nar/13.8.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli K-12 clones that overproduce dam methylase are hypermutable. J Bacteriol. 1981 Jan;145(1):644–646. doi: 10.1128/jb.145.1.644-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Hoess R. H., Fan D. P. Further characterization of a non-essential mutator gene in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):650–660. doi: 10.1128/jb.124.2.650-660.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Herman R. K. Isolation and characterization of mutator strains of Escherichia coli K-12. J Bacteriol. 1975 May;122(2):474–484. doi: 10.1128/jb.122.2.474-484.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Marinus M. G. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982 Apr 29;296(5860):868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Su S. S., Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L. Influence of GATC sequences on Escherichia coli DNA mismatch repair in vitro. J Bacteriol. 1987 Mar;169(3):1254–1259. doi: 10.1128/jb.169.3.1254-1259.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Längle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987 Apr;6(4):1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974 May 15;85(2):309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975 Apr;28(1):15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Poteete A., Arraj J. A. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984 Apr;28(1):123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- McGraw B. R., Marinus M. G. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178(2):309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Pang P. P., Lundberg A. S., Walker G. C. Identification and characterization of the mutL and mutS gene products of Salmonella typhimurium LT2. J Bacteriol. 1985 Sep;163(3):1007–1015. doi: 10.1128/jb.163.3.1007-1015.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P. P., Tsen S. D., Lundberg A. S., Walker G. C. The mutH, mutL, mutS, and uvrD genes of Salmonella typhimurium LT2. Cold Spring Harb Symp Quant Biol. 1984;49:597–602. doi: 10.1101/sqb.1984.049.01.067. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Rewinski C., Marinus M. G. Mutation spectrum in Escherichia coli DNA mismatch repair deficient (mutH) strain. Nucleic Acids Res. 1987 Oct 26;15(20):8205–8215. doi: 10.1093/nar/15.20.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen A. 2-Aminopurine. Mutat Res. 1980 Jan;75(1):1–47. doi: 10.1016/0165-1110(80)90026-3. [DOI] [PubMed] [Google Scholar]
- Rydberg B. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 1978 Oct;52(1):11–24. doi: 10.1016/0027-5107(78)90091-x. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S., Marinus M. G. Direct role of the Escherichia coli Dam DNA methyltransferase in methylation-directed mismatch repair. J Bacteriol. 1986 Mar;165(3):896–900. doi: 10.1128/jb.165.3.896-900.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanabruch W. G., Rein R. P., Behlau I., Walker G. C. Mutagenesis, by methylating and ethylating agents, in mutH, mutL, mutS, and uvrD mutants of Salmonella typhimurium LT2. J Bacteriol. 1983 Jan;153(1):33–44. doi: 10.1128/jb.153.1.33-44.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. C., Smith K. C. Inviability of dam recA and dam recB cells of Escherichia coli is correlated with their inability to repair DNA double-strand breaks produced by mismatch repair. J Bacteriol. 1986 Mar;165(3):1023–1025. doi: 10.1128/jb.165.3.1023-1025.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. M., Goodman M. F. On the molecular basis of transition mutations: frequencies of forming 2-aminopurine.cytosine and adenine.cytosine base mispairs in vitro. Proc Natl Acad Sci U S A. 1981 May;78(5):2864–2868. doi: 10.1073/pnas.78.5.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]
- Whitcome P., Fry K., Salser W. The use of ribosubstitution techniques for determining DNA sequences. Methods Enzymol. 1974;29:295–321. [PubMed] [Google Scholar]



