Abstract
In some organisms longevity, growth, and developmental rate are improved when they are maintained on a light/dark cycle, the period of which “resonates” optimally with the period of the endogenous circadian clock. However, to our knowledge no studies have demonstrated that reproductive fitness per se is improved by resonance between the endogenous clock and the environmental cycle. We tested the adaptive significance of circadian programming by measuring the relative fitness under competition between various strains of cyanobacteria expressing different circadian periods. Strains that had a circadian period similar to that of the light/dark cycle were favored under competition in a manner that indicates the action of soft selection.
Circadian (daily) rhythms are ubiquitous in eukaryotes and are also found in eubacteria among cyanobacteria (1, 2). This ubiquity suggests that these endogenous circadian programs enhance reproductive fitness, but, to our knowledge, there has been no rigorous test of this postulation. Previous investigations have studied related parameters such as longevity, growth, and developmental rate. In some cases, these parameters are improved when organisms are maintained on light/dark (LD) cycles, the periods of which “resonate” optimally with the free-running period (FRP) of the endogenous circadian clock (3–10). For example, the longevity of insects was reported to be maximal in LD cycles that most closely matched that of the FRP (3–5). However, the primary study (3) that purported to demonstrate this effect could not be repeated, even in the laboratory from which the report originated (T. Page, personal communication). The developmental rate of some insect species is most rapid in LD cycles with a period of 24 h (6), but in other species, there is little effect of such protocols (11). A number of studies of some higher plants showed that vegetative growth rate is optimal under LD cycles whose period corresponds with that of the FRP (7–10). In particular, constant light (LL) is deleterious (9, 10). However, whereas some plant species (especially tomato) are sensitive to this inhibition by LL, many other plant species are essentially unaffected (9).
Nevertheless, the parameters of longevity, growth, and developmental rate do not directly assess fitness. An appropriate way to determine the adaptive significance of circadian programs would be to examine the reproductive fitness of mutants whose circadian clockwork is altered. However, FRP or clock-null mutations of circadian clock genes in Drosophila (per) or Neurospora (frq) do not obviously impair reproduction, growth, or development under laboratory conditions (ref. 11; J.C. Dunlap, personal communication). [An exception is a long-period allele of per (perl) that has a pleiotropic effect on the development of Drosophila such that mutant flies develop significantly slower than wild-type flies but that confers no other obvious deficiency (11). Furthermore, mammals rendered arrhythmic by ablation of the suprachiasmatic nuclei do not suffer impairment of reproductive fitness under laboratory conditions, although one study in a simulated field condition suggested that arrhythmic ground squirrels are more susceptible to predation (12).
Therefore, we are unaware of any study to date that has rigorously demonstrated that reproductive fitness per se is improved by resonance between the endogenous clock and the environmental cycle. We addressed the adaptive significance of circadian programs in competition experiments by using the asexual cyanobacterium Synechococcus sp. strain PCC 7942. This cyanobacterium is not known to conjugate under laboratory conditions, and we have derived strains of it that exhibit different FRPs (13). For asexual microbes, differential growth of one strain under competition with other strains is a good measure of reproductive fitness (14). This method is easily adapted to studying parameters that affect fitness of this cyanobacterium. We found conclusive evidence that strains with a circadian period similar to that of the LD cycle were favored under competition in a manner that indicates the action of soft selection (15).
MATERIALS AND METHODS
Strains.
The strains were selected on the basis of equivalent growth rates in LL (13) and stability of FRP in all phases of growth in a liquid or solid medium (data not shown). Strains that had wild-type FRPs (≈25 h at 30°C) were AMC149 (spectinomycin resistance) and AMC343 (chloramphenicol resistance), which were genetically engineered from Synechococcus sp. strain PCC7942 by targeting a luxAB reporter under the control of the psbAI promoter to a neutral site (ref. 16; unpublished data). From AMC149, SP22 (FRP ≅ 23 h) and P28 (FRP ≅ 30 h) were derived by ethyl methanesulfonate mutagenesis; they are spectinomycin resistant (13). SP22 and P28 have different point mutations in the kaiC gene that result in single amino acid changes (17). The plating efficiency of the various strains ranged from approximately 40 to 55% but was consistent within each strain for plating from either logarithmic or stationary liquid cultures onto solid medium.
Culture Conditions.
BG-11 medium (ref. 18, modified as in ref. 19) was used for both liquid and solid media. Cell number (by Coulter Counter) or OD750 was measured for either mixed or pure culture growth (early stationary phase of ca. OD750 = 1.0). For the experiments depicted in Fig. 4, different strains were plated on three dishes and incubated in LL for 2–3 days and then under LD 11:11, LD 12:12, or LD 15:15 for four or five cycles. All strains were then placed into LL at the same phase (subjective dawn), and their circadian FRP, phase, and waveform were determined with a charge-coupled device (CCD) camera/turntable apparatus (ref. 13) or with a TopCount scintillation counter (Packard).
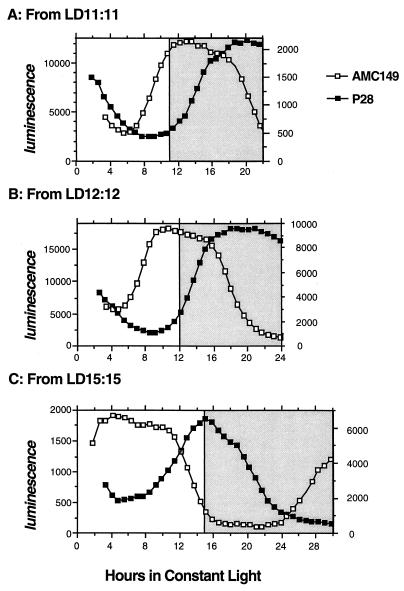
Figure 4.
Phase relationships of the rhythms of wild type (AMC149) and P28 after entrainment to LD cycles. These are composites showing the average trace for 60–200 colonies. Colonies were entrained to LD 11:11, LD 12:12 or LD 15:15 for four to five cycles and then transferred to LL at time 0; their circadian rhythms were monitored for the first cycle in LL. Data shown are after entrainment to LD 11:11 (A), LD 12:12 (B), and LD 15:15 (C). All data are from LL; shaded areas are the extrapolated night phases of the prior LD cycles. Ordinates are the luminescence intensities (left, for AMC149; right, for P28).
Competition Experiments.
For batch cultures (see Figs. 2–4), incubations were performed at 30°C with aeration under white fluorescent light (90–100 μM⋅m−2⋅s−1, measured with a planar light detector). When the cultures reached an OD750 of approximately 1.0, two strains were diluted about 1:1000 and mixed at approximately equal densities. The mixed population was immediately plated so that the initial composition of the population could be measured. For mixed cultures to be placed in LD cycles, the onset of the light phase of the cycle was initiated immediately after this first plating. The cultures were diluted every 8 days. The number of generations was estimated from counting cell numbers with the Coulter Counter.
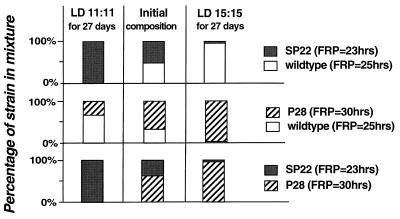
Figure 2.
Growth of various cyanobacterial strains in competition with each other in batch cultures. Pairs of strains were mixed together and an aliquot was plated to form single colonies to determine the initial composition of the culture. The strains were grown for 27 days in batch culture under either a 22-h LD cycle (LD 11:11) or a 30-h LD cycle (LD 15:15), and then another aliquot of the mixed culture was plated to determine the 27-day composition of the culture. Each horizontal sequence shows the results for the pairs of strains listed on the right. The composition of the cultures was determined by observing the luminescence patterns of the colonies and counting the number of colonies on each plate exhibiting each of the strain-specific phenotypes by using a CCD camera/turntable apparatus (13). In the experiments depicted in this figure, wild type was strain AMC149. The experiment was repeated three times with essentially the same outcomes.
For competition experiments (Table 1) conducted in the turbidostat (20), cultures were grown to an OD750 of approximately 0.4, diluted to an OD750 of 0.2, and then mixed in equal volumes. A day 0 sample was taken to confirm an equal frequency of the two phenotypes. For cultures in an LD cycle, the onset of the light phase started at the onset of turbidostat culturing. All turbidostat experiments were conducted with white light at 100 μM⋅m−2⋅s−1 (measured with a spherical light detector), and cultures were bubbled vigorously with air. Because the turbidostat maintains a constant cell density (in these experiments, OD750 ≅ 0.4), generation number was estimated by changes in culture volume.
Table 1.
Turbidostat data
| Days, n | LD 12:12
|
LD 15:15
|
Generations, n | ||||
|---|---|---|---|---|---|---|---|
| % wt | % P28 | n | % wt | % P28 | n | ||
| 0 | 52 | 48 | 87 | 52 | 48 | 87 | |
| 15 | 96 | 4 | 77 | 5 | 95 | 84 | 5.4 |
| 19 | 100 | 0 | 85 | 4 | 96 | 83 | 6.8 |
Determination of Composition of Populations under Competition.
At regular intervals during growth, aliquots of cultures were plated during an illuminated phase of the LD cycle, and the plates were grown in LL. Composition of populations from plates was scored by two different methods. First, for competitions between strains that had the same antibiotic resistances (see Fig. 2), the circadian phenotype (FRP) of each colony on plates in LL was assessed by either a CCD camera/turntable apparatus (13), in which case three plates were measured for each time, or with a TopCount, in which case at least one 96-well plate (see n values in Table 1) was measured for each time. The number of colonies of either phenotype reflects the fraction of that strain in the population.
To assess time points more frequently, we resorted to a second method of scoring that took advantage of differential antibiotic resistances of the mutant strains (spectinomycin resistant) and the wild-type AMC343 (chloramphenicol resistant) to measure the kinetics of selection (see Fig. 3). Each mixed culture was plated onto six Petri dishes with three dishes containing spectinomycin (20 μg/μl) and the other three containing chloramphenicol (4 μg/μl), and the number of colonies on each plate was counted manually or by a CCD camera apparatus (Alpha Innotech, San Leandro, CA). For both scoring methods, only plates with total colony numbers ranging between 40 and 2000 were used for data analysis.
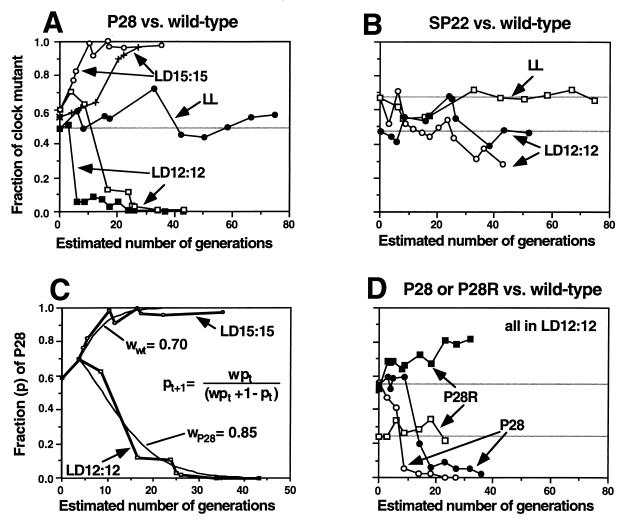
Figure 3.
Kinetics of competition between wild-type and mutant/rescued strains (batch cultures). (A) Competition between wild type (AMC343) and P28 in LD 12:12, LD 15:15, and LL plotted as a function of estimated number of generations. The results for two experiments (of five independent experiments) are shown for each LD treatment. Ordinate, Percentage of colonies in the population that are P28. (B) Competition between wild type (AMC343) and SP22 in LD 12:12 and LL plotted as in A. The results for two independent LD experiments are shown. Ordinate, Percentage of colonies in the population that are SP22. (C) Comparison of the open-symbol raw data from A (points and heavy lines) with the results of modeling (thin lines) by using the equation shown, where w = relative fitness [wwt for AMC343 (wild type), wP28 for P28] and p = fraction of a given strain in the mixed population [pt for generation (t), pt+1 for generation (t + 1)]. (D) Competition between wild type (AMC343) and either P28 (FRP ≅ 30 h) or P28R (FRP ≅ 25 h) in LD 12:12. Two independent experiments are shown (open and closed symbols). Ordinate, Percentage of colonies in the population that are P28 or P28R. For all panels, the horizontal lines are the predictions if the initial population compositions are maintained.
Construction of P28R from P28.
The mutant kaiC allele was replaced with the wild-type allele in a two-step “hit and run” procedure, and the result was the rescued P28R. P28 cells (10 ml) were washed with 10 mM NaCl and resuspended in 1 ml BG-11 with ≈200 ng/ml pAM1736 plasmid, which has a wild-type kaiC sequence inserted into the pRL278 vector (pRL278 carries a kanamycin resistance gene and the sacB gene; ref. 21). Cells were incubated with pAM1736 at 30°C for 3–4 h in darkness with gentle shaking. Then, 150 μl of this culture was plated on BG-11 with kanamycin (20 μg/ml) to select for integration of the plasmid by single crossover homologous recombination. A few randomly chosen colonies were incubated in liquid BG-11 without kanamycin and then plated on BG-11 that contained 5% sucrose but no kanamycin to select for “loopout” of the plasmid, the reverse of the integration event. Colonies that were sensitive to kanamycin, thereby confirming the loopout, were screened; of about 100 colonies whose FRP was rescued from approximately 30 to 25 h, one was used to inoculate the cultures used in the experiment depicted in Fig. 3D.
RESULTS AND DISCUSSION
Competition Detects Fitness Differences That Cannot Be Measured in Pure Cultures.
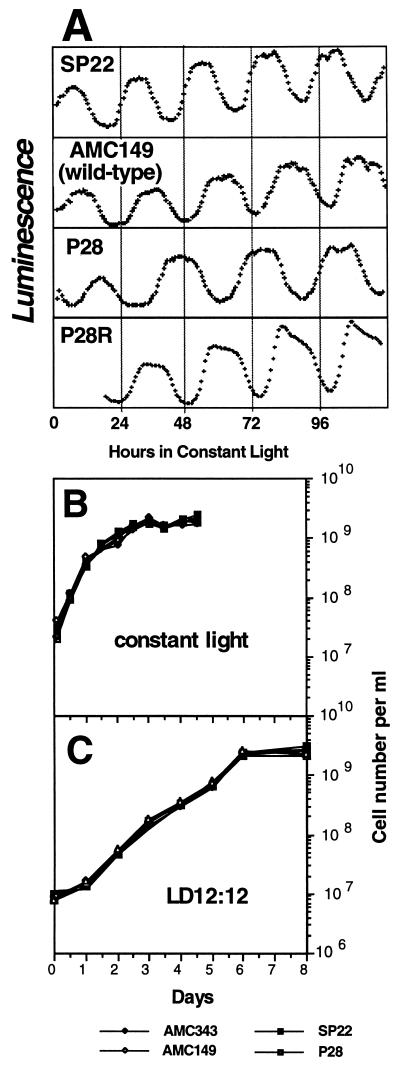
The phenotypes of the strains we tested are illustrated in Fig. 1; A shows that the FRPs at 30°C in LL of the strains are ≈23 h (SP22), 25 h (wild-type strains AMC149 and AMC343), and 30 h (P28). SP22 and P28 each harbor a missense mutation in a cyanobacterial clock gene, kaiC (17), and were chosen for the present study because their FRPs are consistent in both logarithmic and stationary phase growth cultures. The growth rates of the various strains in pure culture are indistinguishable in both LL (Fig. 1B) and a 24-h LD 12:12 cycle (Fig. 1C). Results similar to those shown in Fig. 1C were also found for LD 11:11 and LD 15:15 (data not shown).
Figure 1.
Characteristics of the strains used in the competition experiments. (A) The circadian phenotypes of different strains as determined with the CCD camera/turntable device described (13). All strains have a luciferase construct that reports the promoter activity of the psbAI gene (16, 22). AMC343 has almost the same phenotype as AMC149; therefore, it is not shown here. FRPs of the different strains at 30°C are approximately: 23 h (SP22), 25 h (AMC149 and AMC343), and 30 h (P28). P28R is the P28 strain that has been genetically rescued to an FRP ≈25 h. (B and C) Growth curves of the individual strains in A. On average, the doubling times for LD were one division per 16.1 h and for LL the doubling times were one division per 6.3 h.
Therefore, there is no apparent difference in growth rate among these strains when grown separately. When each of the strains is mixed with another strain and grown together in competition, however, a pattern emerges that depends on the nature of the LD cycle. We tested two different LD cycles that had equal amounts of light and darkness but the frequency of the LD cycle differed: a 22-h cycle (LD 11:11) and a 30-h cycle (LD 15:15). As illustrated in Fig. 2, when wild type competes against SP22, wild-type cells grow better during 27 days of competition in the 30-h cycle, but the short-period SP22 defeats wild type in the 22-h cycle. When competing against wild-type, the long-period P28 takes over the culture in the 30-h cycle but loses in the 22-h cycle. When P28 competes against SP22, the results are clear: the strain whose period most closely matches that of the LD cycle eliminates the competitor. When the two wild-type strains (AMC149 and AMC343) compete against each other, both strains are maintained in the population for many generations (data not shown).
More detailed studies of the kinetics of competition between wild type and the mutant P28 are shown in Fig. 3A. Wild type out-competes P28 quickly in LD 12:12, whereas P28 defeats wild-type in LD 15:15. When these same combinations are grown together in LL, however, both strains are maintained, indicating that, in the absence of a LD cycle, the fitness of the mutant strain is approximately equal to that of wild type. To test whether the results obtained with batch cultures could be replicated under other conditions, we also competed wild type against P28 in a turbidostat apparatus that allows the maintenance of constant cell density and nutrients (20); in the turbidostat, wild type also defeats P28 in LD 12:12 but loses to P28 in LD 15:15 (Table 1). Because the turbidostat cultures were maintained in midlogarithmic phase, these data indicate that competition can occur in continuously growing cultures. Another component of the competition that may be important in batch cultures, however, is the lag in growth recovery after dilution into fresh medium.
Fig. 3B illustrates the results of two independent competition experiments between wild type and SP22 in LD 12:12. In one case, wild type appears to out-compete SP22 (but less strongly than in the combinations shown in Fig. 3A), whereas in the other experiment both strains are maintained. These results are consistent with a small selective difference between the FRPs of wild type (≈25 h) vs. SP22 (≈23 h) as compared with the period of the LD cycle (24 h). Again, both strains coexist in LL. The phenomena we observed are not caused by chance or initial conditions because the competition can be started at different initial frequencies of the strains and the same final results are obtained (data not shown).
Estimation of Selective Advantage Conferred by Circadian Resonance.
To obtain an estimate of the relative fitness of P28 vs. wild type under LD12:12 as well as LD15:15, we used a simple model that assumes (i) the selective pressure to be constant and (ii) a lag in the initiation of selection (Fig. 3C). Although this simple model does not model the data perfectly, it suggests to a first approximation that the relative fitness of the less successful strain could be as low as 0.7–0.8 (relative fitness of wild type ≅ 0.7 in LD 15:15, relative fitness of P28 ≅ 0.85 in LD 12:12). Because the growth rates of the strains in pure culture do not appear to be different by a factor of 20–30% (Fig. 1 B and C), the modeling result depicted in Fig. 3C indicates that we are observing a case of soft selection where the poorer fitness of inferior genotypes is most obvious under competition (15). We cannot rule out that small, presently unmeasurable, differences exist between the growth rates of these strains in pure culture, but soft selection seems to be the predominant mechanism responsible for the strong selection under competition.
Selective Advantage Is Not Caused by Secondary Mutations.
The results shown in Figs. 2 and 3 A–C indicate that the strain whose FRP resonates nearest to the period of the environmental cycle is the most fit in these competition experiments. Because the mutants P28 and SP22 can defeat wild type in LD cycles whose periods are similar to their FRPs, the differential effects we observed are likely to be caused by the differences in the circadian clock and are unlikely to be because of an unrelated mutation that is deleterious for growth. To test that conclusion directly, we constructed a strain of P28 in which the FRP was rescued to that of wild type, called P28R (FRP shown in Fig. 1A). The mutation that is responsible for the long period phenotype of P28 is known to be in the kaiC gene (17); therefore, we replaced the mutated gene with wild-type kaiC. With the exception of the kaiC gene, P28 and P28R are genetically identical. Fig. 3D shows that, when P28R competes against wild type in LD 12:12, it is no longer defeated but can coexist with wild type. This result excludes the interpretation that the differential growth characteristics we report here are because of a secondary mutation that is unrelated to the clock.
Phase Relationships Are Different Between Resonating and Nonresonating Combinations.
We do not presently know the physiological mechanism by which one strain out-competes the other. Several factors could be responsible, e.g., competition for limiting resources such as light, nutrients, and carbon dioxide. It is also possible that strains of cyanobacteria rhythmically secrete diffusible factors that inhibit the growth of other cyanobacterial strains. Are the results depicted in Figs. 2 and 3 because of a failure of the cyanobacterial clock to entrain to LD cycles that are significantly different from their FRP? Apparently not. When the phases of the luminescence rhythms (reporting psbAI promoter activity) of individual colonies are compared after growth in LD 11:11, 12:12, or 15:15, all colonies within each strain type are in phase with each other. Furthermore, measurements of the phase response curves of wild type, SP22, and P28 to 12-h dark pulses indicate that each of these strains would be able to entrain to LD cycles that are ±10 h or more from their FRP (data not shown).
The most likely explanation for the differences in selective advantage is that the phase relationships between biological rhythms and environmental cycles are predictably altered in different combinations of FRP vs. the period of the LD cycle (23). Indeed, the phase relationships between the luminescence rhythms and the LD cycles are different among the strain types (Fig. 4). The optimal combinations of FRP/LD period (e.g., wild type on LD 12:12 and P28 on LD 15:15) are correlated with a phase relationship of rhythmic psbAI promoter activity being low in the early day and peaking near dusk (Fig. 4). This result confirms that the phase relationship of the circadian timekeeper is altered when entrained to different LD cycles; however, note that the rhythm of psbAI promoter activity might not be the rhythmic output that is most directly related to the selection.
Whatever specific processes are responsible for the selective advantage we have observed, our experiments clearly indicate that having a circadian clock with an FRP similar to that of the environmental cycle is adaptive for the cyanobacterium Synechococcus. Our results are not because of noncircadian mutations in our strains, because the mutant strains can defeat wild type under LD cycles that resonate optimally with the mutant FRPs (Figs. 2 and 3), and rescuing the circadian phenotype of the clock mutant P28 also recovers the competitive ability of the strain in LD 12:12 (Fig. 3D). Moreover, the competition results are the same under competition between strains with the same (Fig. 2) or different (Fig. 3) antibiotic resistances. Therefore, our results are not an artifact of different selective markers.
The circadian clock mutants that we studied were not impaired in growth or reproduction when grown in pure culture; therefore, growth in competition is a very sensitive method by which to assess reproductive fitness conferred by a circadian program. Based on our data, we conclude that the circadian pacemaker in cyanobacteria confers a significant competitive advantage in LD cycles when the FRP of the clock resonates with the environmental cycle so as to achieve an optimal phase relationship between the LD cycle and the internal timekeeper.
Acknowledgments
We thank D. Taylor, D. McCauley, R. Lenski, C. dePamphilis, C. P. Kyriacou, T. Page, and S. Daan for helpful comments. We thank Dr. Masahiro Ishiura for providing the plasmid used to construct AMC343 (PpsbAI∷luxAB linked to chloramphenicol resistance) and Jeff Plautz for assistance with the analysis of TopCount data. We dedicate this paper to Colin S. Pittendrigh, a pioneer of circadian biology whose first concern was to understand the functional significance of biological clocks. This work was supported by grants from the National Institute of Mental Health (MH 01179 to C.H.J.) and the National Science Foundation (MCB-9633267 to C.J. and MCB-9513367 to S.G.).
Footnotes
References
- 1.Edmunds L N. Cellular and Molecular Bases of Biological Clocks. New York: Springer; 1988. [Google Scholar]
- 2.Johnson C H, Golden S S, Ishiura M, Kondo T. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 3.Pittendrigh C S, Minis D H. Proc Natl Acad Sci USA. 1972;69:1537–1539. doi: 10.1073/pnas.69.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aschoff J, v. Saint Paul U, Wever R. Naturwissenschaften. 1971;58:574. doi: 10.1007/BF00598736. [DOI] [PubMed] [Google Scholar]
- 5.St. Paul U, Aschoff J. J Comp Physiol. 1978;127:191–195. [Google Scholar]
- 6.Saunders D S. Proc Natl Acad Sci USA. 1972;69:2738–2740. doi: 10.1073/pnas.69.9.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Withrow A P, Withrow R B. Plant Physiol. 1949;24:657–663. doi: 10.1104/pp.24.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Highkin H R, Hanson J B. Plant Physiol. 1954;29:301–302. doi: 10.1104/pp.29.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillman W S. Plant Physiol. 1956;43:89–95. [Google Scholar]
- 10.Went F W. Cold Spring Harbor Symp Quant Biol. 1960;25:221–230. doi: 10.1101/sqb.1960.025.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Kyriacou C P, Oldroyd M, Wood J, Sharp M, Hill M. Heredity. 1990;64:395–401. doi: 10.1038/hdy.1990.50. [DOI] [PubMed] [Google Scholar]
- 12.DeCoursey P J, Krulas J R, Mele G, Holley D C. Physiol Behav. 1997;62:1099–1108. doi: 10.1016/s0031-9384(97)00263-1. [DOI] [PubMed] [Google Scholar]
- 13.Kondo T, Tsinoremas N F, Golden S S, Johnson C H, Kutsuna S, Ishiura M. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 14.Lenski R E, Travisano M. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futuyma D J. Evolutionary Biology. 3rd Ed. Sunderland, MA: Sinauer; 1998. p. 386. [Google Scholar]
- 16.Kondo T, Strayer C A, Kulkarni R D, Taylor W, Ishiura M, Golden S S, Johnson C H. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H. & Kondo, T. (1998) Science, in press. [DOI] [PubMed]
- 18.Allen M M. J Phycol. 1968;4:1–4. doi: 10.1111/j.1529-8817.1968.tb04667.x. [DOI] [PubMed] [Google Scholar]
- 19.Bustos S A, Golden S S. J Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustos S A, Golden S S. Mol Gen Genet. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]
- 21.Cai Y P, Wolk C P. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Golden S S, Kondo T, Ishiura M, Johnson C H. J Bacteriol. 1995;177:2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aschoff J. Handbook of Behavioral Neurobiology. Vol. 4. New York: Plenum; 1981. pp. 81–93. [Google Scholar]