Abstract
Mutations of DNA mismatch repair genes, including the hMLH1 gene, have been linked to human colon and other cancers in which defective DNA repair is evidenced by the associated instability of DNA microsatellite sequences (MSI). Germ-line hMLH1 mutations are causally associated with inherited MSI colon cancer, and somatic mutations are causally associated with sporadic MSI colon cancer. Previously however, we demonstrated that in many sporadic MSI colon cancers hMLH1 and all other DNA mismatch repair genes are wild type. To investigate this class of tumors further, we examined a group of MSI cancer cell lines, most of which were documented as established from antecedent MSI-positive malignant tumors. In five of six such cases we found that hMLH1 protein was absent, even though hMLH1-coding sequences were wild type. In each such case, absence of hMLH1 protein was associated with the methylation of the hMLH1 gene promoter. Furthermore, in each case, treatment with the demethylating agent 5-azacytidine induced expression of the absent hMLH1 protein. Moreover, in single cell clones, hMLH1 expression could be turned on, off, and on again by 5-azacytidine exposure, washout, and reexposure. This epigenetic inactivation of hMLH1 additionally accounted for the silencing of both maternal and paternal tumor hMLH1 alleles, both of which could be reactivated by 5-azacytidine. In summary, substantial numbers of human MSI cancers appear to arise by hMLH1 silencing via an epigenetic mechanism that can inactivate both of the hMLH1 alleles. Promoter methylation is intimately associated with this epigenetic silencing mechanism.
Germ-line defects in DNA mismatch repair (MMR) genes account for the inherited familial cancer syndrome of hereditary nonpolyposis colon cancers in which affected individuals show accelerated development of cancers of the proximal colon, endometrium, and stomach (1–5). These cancers typically demonstrate inactivation of the residual wild-type MMR allele inherited opposite the germ-line mutant (1–6), absence of DNA MMR activity in in vitro assays (7, 8), and acquisition of an in vivo mutator phenotype showing up to 1,000-fold increased gene mutation rates (9, 10). Additionally, these cancers display an associated instability of genomic microsatellite sequences (MSI) (1–5). MSI is similarly found in approximately 15–20% of sporadic colon cancers that arise in individuals without any family history of colon cancer (1–5). Similarly to hereditary nonpolyposis colon cancer-associated colon cancers, sporadic MSI colon cancers arise predominantly in the proximal colon and show a high rate of frameshift mutations at a mutational hotspot in the transforming growth factor-β type II receptor tumor suppressor gene (1, 11). Familial and sporadic MSI colon cancers thus appear to share a common carcinogenic pathway. In this regard, previous studies from our group established that MMR gene inactivation via somatic mutation was the cause of some cases of sporadic MSI colon cancers (12). However, unexpectedly, in many sporadic MSI colon cancers, MMR genes were found to remain wild type (12). MMR-coding sequences have similarly been reported to be wild type in most sporadic MSI endometrial cancers (13). Thus, a mechanism that induces MSI independently of MMR gene mutation appears to account for a significant number of human MSI cancers. Intriguingly, methylation of the hMLH1 promoter region has recently been described in some MSI tumors (14).
In this study we focused on further elucidating the mechanism that results in MSI in sporadic cancers bearing apparently wild-type MMR genes. We report here that, in five sporadic MSI cancers (four colon and one endometrial) in which MMR genes are wild type, MSI arises via a new epigenetic mechanism that inactivates MMR function by silencing transcription of the wild-type hMLH1 MMR gene. Epigenetic inactivation of hMLH1 is biallelic, is associated with methylation of the hMLH1 promoter, and is reversed by the demethylating agent 5-azacytidine. Moreover, hMLH1 inactivation is reestablished after withdrawal of the demethylating agent, suggesting that an underlying active process maintains the inactivated methylated state.
MATERIALS AND METHODS
Cell Culture.
Cells were cultured in MEM supplemented with gentamycin, l-glutamine, nonessential amino acids, and sodium selenite. AN3CA growth medium was supplemented with 10% fetal bovine serum and sodium pyruvate. For HCT116, Vaco5, and Vaco6, growth medium was supplemented with 8% calf serum. For SW480, RKO, Vaco432, Vaco457 and Vaco703, growth medium was supplemented with 2% fetal bovine serum (HyClone), 10 mg/liter bovine insulin, 2 mg/liter human transferin, and 1 mg/liter hydrocortisone. Monolayer cultures were grown at 37°C in a 5% CO2 atmosphere. The Vaco cell lines were established from colon cancers as described (11, 15). HCT116, AN3CA, and SW480 were obtained from the American Type Culture Collection, and RKO was a generous gift from M. Brattain (University of Texas Health Science Center, San Antonio, TX).
5-Azacytidine Treatment.
Cells were seeded at 105 cells per T75 flask on day 0. The cultures were treated for 24 h on day 2 and day 5 with 5-azacytidine at 1–3 μg/ml as individually tolerated. The medium was changed 24 h after addition of the 5-azacytidine (i.e., on day 3 and day 6). On day 8 the cells were harvested for analysis of the methylation status of the hMLH1 promoter region and of production of hMLH1 protein. Additionally, in some experiments on day 8 cells were either reseeded for continued analysis of hMLH1 expression in bulk culture or subcloned by limiting dilution on 96-well culture plates.
Evaluation of Methylation Status of the hMLH1 Promoter Region.
The methylation status of the hMLH1 promoter was evaluated by a PCR-based assay according to the procedure detailed by Kane et al. (14). Briefly, genomic DNA samples were digested with no enzyme, HpaII, or MspI, and then the 603-bp region upstream of the hMLH1 gene, which contains four HpaII sites, was amplified by PCR. The amplification products were then visualized with ethidium bromide after agarose gel electrophoresis.
Western Analysis.
Approximately 107 cells were lysed in cell lysis buffer [50 mM Tris⋅HCl (pH 7.4)/1 mM EGTA/1% Nonidet P-40/0.25% sodium deoxycholate/150 mM NaCl] and were subjected to electrophoresis through an SDS polyacrylamide gel. The resultant proteins were then transferred to a poly(vinylidene difluoride) nylon membrane, which was probed with 2 μg/ml anti-human MLH1 monoclonal antibody (PharMingen). Immune complexes were visualized by using an enhanced chemiluminescence reagent (Amersham) after incubation with horseradish peroxidase-coupled secondary antibody (Calbiochem). The membrane was then deprobed and reprobed with 300 ng/ml anti-human actin monoclonal antibody (Amersham).
Amplification and Sequencing of hMLH1 Genomic and cDNA.
Genomic DNA was obtained by lysing cells in proteinase K buffer [10 mM Tris (pH 7.4)/10 mM EDTA/0.4% SDS/150 mM NaCl/100 μg/ml proteinase K] and extracting with phenol/chloroform/isoamyl alcohol (25:24:1). Genomic DNA was amplified in a thermocycler for 38 cycles by using the following protocol: cycle 1, 95°C × 5 min; subsequent cycles, 95°C × 45 s, 55°C × 30 s, and 72°C × 30 s; final cycle, 72°C × 5 min and hold at 4°C. Each 30-μl reaction contained 100 ng genomic DNA, 0.125 mM 2′-deoxynucleoside 5′-triphosphate, 300 ng forward primer, 300 ng reverse primer, and 2.5 units Perkin–Elmer Amplitaq Gold polymerase.
Cells were treated with guanidine isothiocyanate, and RNA was isolated by sedimentation through cesium chloride as described (16). The cDNA for hMLH1 was amplified by using the Titan RT-PCR system (Boehringer Mannheim). Reverse transcription was carried out for 30 min at 50°C and then by 38 cycles of PCR (cycle 1, 95°C × 2 min; subsequent cycles, 95°C × 45 s, 54°C × 45 s, and 68°C × 1 min; final cycle, 68°C × 7 min and hold at 4°C).
The primer sequences were chosen by using macvector 5.0./2 (International Biotechnology, New Haven, CT) software and the following GenBank hMLH1 sequence accession numbers: U83845 for the promoter, U07418 for the cDNA, and U40967 for exon 8. A 217-bp genomic fragment encompassing hMLH1 codon 219 was amplified with primers MLHEX8F1 (5′-CTCAGCCATGAGACAATAAATCC) and MLHEX8R1 (5′-GGTTCCCAAATAATGTGATGG). An hMLH1 cDNA fragment encompassing codon 219 and spanning exons 7–9 was amplified with primers MLH564S (5′-CAGGTATTCAGTACACAATGCAGGC) and MLH823AS (5′-CTACCAGACGATGGTTGATGAAGAG). The putative hMLH1 promoter region extending from 765 bp 5′ to 57 bp 3′ of the ATG was amplified with primers MLHF5 (5′-CACGAGCAGTCTCTCTTCAGGAGT) and MLHR5 (5′-GATGCGGTTCACCACTGTCTCG). Sequencing of the codon 219 polymorphism was done with primer MLHEX8F1 in genomic DNA samples and primer MLH564S in cDNA samples. Sequencing primers for the hMLH1 promoter region are available from the authors by request.
RESULTS
Loss of MLH1 Protein Expression and Functional Activity in Sporadic MSI Cancers.
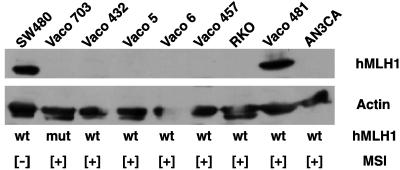
We initially investigated five cell lines established from sporadic MSI colon cancer tumors that we had previously reported bore only wild-type MMR genes (Vaco432, Vaco5, Vaco6, RKO, and Vaco481) (12). Fig. 1 depicts our Western blot analysis that demonstrates absence of hMLH1 protein expression in four of these five cell lines (Vaco432, Vaco5, Vaco6, and RKO). Furthermore, the western blot shows that no hMLH1 protein was expressed in a sporadic MSI endometrial cancer, AN3CA, which previously was shown to possess wild-type MMR genes but to lack hMLH1 transcript (17). Finally, hMLH1 protein was also absent in the Vaco703 colon cancer, which we previously reported carries an inactivating somatic mutation of the hMLH1 gene (12), and in Vaco457, a hereditary nonpolyposis colon cancer-derived colon cancer. Expression of hMLH1 protein was, however, readily detected in the non-MSI colon cancer SW480. Northern analysis confirmed that, in sporadic MSI cancers with wild-type hMLH1-coding sequences, those lacking hMLH1 protein expression also lacked hMLH1 transcript expression. Western blots demonstrated continued expression of hMSH2 protein in Vaco5, Vaco6, Vaco432, RKO, and AN3CA. This suggested that in all of these cases the expression of the wild-type hMLH1 alleles had been specifically silenced.
Figure 1.
hMLH1 protein expression in MSI cancers. Shown is a Western blot of hMLH1 protein expression. The presence (+) or absence (−) of MSI and the status of the coding region of the hMLH1 gene (wt, wild type; mut, mutant) are denoted below each lane. The Western analysis was performed as described in Materials and Methods.
To characterize the functional consequences of the observed loss of detectable hMLH1 protein, MMR activity was assayed in nuclear extracts derived from Vaco6, RKO, and AN3CA. In vitro experiments, to be described elsewhere, indicated that Vaco6, RKO, and AN3CA have an MMR defect caused by an MutLα deficiency similar to that observed previously with cisplatin- and adriamycin-resistant ovarian tumor cells (18). Functional repair activity could be restored by addition of hMutLα, the heterodimeric protein complex of hMLH1 and hPMS2, to each of these extracts (19). Thus, the absence of hMLH1 protein expression in these cancers is the direct cause of their MSI phenotype.
Loss of hMLH1 Expression Is Associated with Methylation of the hMLH1 Gene Promoter.
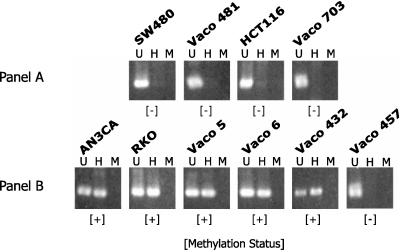
To establish the mechanism of loss of hMLH1 gene expression in AN3CA, RKO, Vaco5, Vaco6, and Vaco432, we first sequenced the hMLH1 promoter for evidence of mutations. No mutations were found in any of these cell lines within 685 bp 5′ of the hMLH1 initiator ATG. We next examined the hMLH1 promoter region in these tumors for evidence of methylation at four HpaII sites located between 341 and 567 bp 5′ of the hMLH1 ATG (14). Methylation was detected by the ability to recover and amplify an intact hMLH1 promoter fragment after digestion of genomic DNA with the methylation-sensitive restriction enzyme HpaII. As shown in Fig. 2B, the five sporadic MSI cancers that displayed loss of hMLH1 expression also have methylation of the hMLH1 promoter (AN3CA, RKO, Vaco5, Vaco6, and Vaco432).
Figure 2.
Analysis of methylation status of the hMLH1 promoter. Shown is the product resulting from PCR amplification of the hMLH1 promoter region before or after digestion. U, Undigested; H, digested with HpaII; M, digested with MspI. The methylation status (+, methylated; −, unmethylated) of the hMLH1 promoter is designated below each sample.
In contrast to hMLH1 promoter methylation in MSI cancers with wild-type MMR genes, the hMLH1 promoter was found to be unmethylated in two MSI cancers in which hMLH1 protein was inactivated by mutation (HCT116 and Vaco703; Fig. 2A), in one MSI cancer in which hMLH1 protein was inactivated by an unknown germ-line defect (Vaco457; Fig. 2B), and in two cancers in which wild-type hMLH1 gene expression was maintained (Vaco481 and SW480). Methylation of the hMLH1 promoter was thus associated with sporadic MSI cancers that lacked MMR gene mutations.
hMLH1 Silencing Is Reversible After Treatment with the Demethylating Agent 5-Azacytidine.
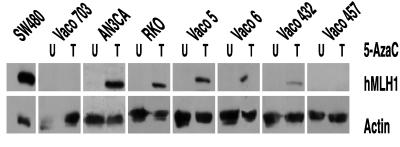
To determine whether hMLH1 promoter methylation could be further linked to the loss of hMLH1 expression, cancer cell lines were treated with the demethylating agent 5-azacytidine. As shown in Fig. 3, 5-azacytidine successfully restored hMLH1 protein expression in each of the five sporadic MSI cancers that lacked hMLH1 expression and that harbored a methylated hMLH1 promoter. In contrast, 5-azacytidine was ineffective in reactivating hMLH1 expression in two MSI cancers in which the hMLH1 promoter was unmethylated and in which hMLH1 inactivation was caused by somatic mutation (Vaco703) or germ-line defect (Vaco457) (Fig. 3). Thus, the presence of hMLH1 promoter methylation was tightly correlated with the ability to reactivate hMLH1 by 5-azacytidine treatment. Moreover, the 5-azacytidine induction of hMLH1 expression in five sporadic MSI cancers repeatedly demonstrated that an epigenetic mechanism must be responsible for silencing of hMLH1 expression.
Figure 3.
Induction of hMLH1 protein expression after 5-azacytidine (AzaC) treatment. The Western analysis was performed as described in Materials and Methods. U, Cells untreated; T, cells treated with 5-azacytidine.
hMLH1 Expression Is Again Silenced After Withdrawal of 5-Azacytidine.
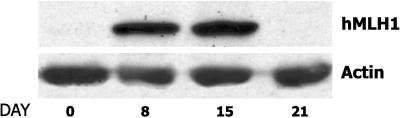
To determine whether hMLH1 gene silencing is actively maintained, we asked whether hMLH1 gene reexpression would persist after washout of the 5-azacytidine-inducing agent. As shown in Fig. 4, 5-azacytidine treatments of AN3CA cells on days 2 and 5 induced abundant hMLH1 protein expression by day 8. However by day 21, 2 weeks after drug washout, hMLH1 protein expression was again undetectable. This result suggested that, after withdrawal of 5-azacytidine, an active process again resilenced expression of the hMLH1 gene.
Figure 4.
Time course of hMLH1 protein expression after 5-azacytidine treatment. AN3CA cells were treated with 5-azacytidine as detailed in Materials and Methods. An untreated sample (day 0), a sample immediately after 5-azacytidine treatment (day 8), and samples 1 week (day 15) and 2 weeks (day 21) after 5-azacytidine treatment were collected for Western analysis to evaluate production of hMLH1 protein.
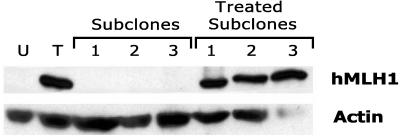
Alternatively, during the 14 days after drug washout the AN3CA culture could be overgrown by a subpopulation of cells resistant to 5-azacytidine. To distinguish between these two possibilities, we reprised the 5-azacytidine treatment of AN3CA with the addition that, after withdrawal of the 5-azacytidine, 22 independent single-cell subclones were immediately established from the AN3CA culture. As shown in Fig. 5, in this repeat study 5-azacytidine again reinduced hMLH1 protein expression in the AN3CA cancer cells by day 8. However, in each of the 22 subclones established from the drug-treated population, washout of the drug again was followed by hMLH1 silencing. To avoid any selection bias favoring subcloning of a putative rapidly growing 5-azacytidine-resistant cell population, we included in this analysis subclones that grew either rapidly or slowly after 5-azacytidine withdrawal. Furthermore, retreating 10 of these subclones with 5-azacytidine in each case resulted in reexpression of the hMLH1 protein (Fig. 5), demonstrating that the subclones indeed were comprised of 5-azacytidine-responsive cells. Expression of hMLH1 in AN3CA could thus be cycled from off, to on, to off, and to on, by 5-azacytidine treatment, washout, and retreatment. These findings clearly establish that inactivation of hMLH1 expression in AN3CA is caused by an epigenetic process. Moreover, these findings suggest that hMLH1 silencing is maintained by an active mechanism that is reasserted after washout of the 5-azacytidine inducing agent.
Figure 5.
Reversible expression of hMLH1 protein in single-cell clones treated with 5-azacytidine. Western analysis of hMLH1 protein expression was performed as described in Materials and Methods and is shown for AN3CA cells untreated (U) or on day 8 of treatment with 5-azacytidine (T). As described in Materials and Methods, subclones 1, 2, and 3 were established from the day 8 treated AN3CA cell population immediately after washout of the drug and were analyzed for continued hMLH1 expression when they had reached confluence. The same three subclones were then retreated with 5-azacytidine and again analyzed (treated subclones). Subclones 1 and 3 are representative normal-growth-rate subclones; whereas subclone 2 is representative of the group of slow-growth-rate subclones.
Epigenetic Inactivation of hMLH1 Is Biallelic.
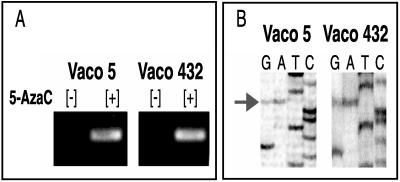
No wild-type hMLH1 protein was detected by Western analysis in the Vaco5, Vaco6, Vaco432, RKO, or AN3CA cell lines, demonstrating in these tumors that both wild-type hMLH1 alleles had been inactivated. In many cancers biallelic inactivation of suppressor genes results from mutation of one allele and then by gene deletion of the remaining allele (1). This two-step process is reflected by loss of heterozygosity of polymorphic markers linked to the suppressor gene locus (1). We looked for such loss of heterozygosity at the hMLH1 locus in three of the cases, Vaco5, Vaco6, and Vaco432, for which matched normal tissue was available. A coding region polymorphism at codon 219 (ATC to GTC) was found to be informative in two cases, Vaco5 and Vaco432 (12). Significantly, retention of both maternal and paternal alleles was detected in each of the Vaco5 and Vaco432 tumors and cell lines. Moreover, after 5-azacytidine treatment, induction of transcripts bearing both maternal and paternal polymorphisms was detected in cDNA amplified from either Vaco5 or Vaco432 (Fig. 6). Thus, in these tumors biallelic inactivation of hMLH1 expression is caused by epigenetic inactivation of both parental copies of the gene. This finding suggests that in these tumors epigenetic silencing of hMLH1 is not a rare chance event but the end result of an active and efficient mechanism that can work in trans. Furthermore, material was available to confirm that the antecedent tumor from which the Vaco432 cell line was established demonstrated both the MSI phenotype as well as methylation of the hMLH1 gene promoter. Thus, epigenetic inactivation of hMLH1 almost certainly was the initiating event in the genesis of the MMR deficiency of this individual’s colonic malignancy.
Figure 6.
Allele-specific analysis of hMLH1 expression after 5-azacytidine treatment in Vaco 5 and Vaco 6. (A) Reverse transcription–PCR analysis of hMLH1 expression in Vaco5 and Vaco432 before (+) and after (−) treatment with 5-azacytidine (AzaC). (B) DNA sequences of hMLH1 cDNAs from Vaco5 and Vaco432 amplified by reverse transcription–PCR after treatment with 5-azacytidine. The arrow points to a coding region polymorphism at codon 219 (ATC to GTC).
DISCUSSION
These findings establish that the epigenetic silencing of the hMLH1 gene promoter is an important event in many sporadic human colon cancers. This epigenetic silencing mechanism is closely associated with methylation of the hMLH1 gene promoter. Moreover, this mechanism was operative in a majority, four of seven, of the sporadic MSI colon cancers in our collection. Additionally, in three of these four cases, we confirmed that the MSI phenotype was present in the antecedent tumors from which the cell lines were established. Thus, the mechanism accounting for MSI in the cell lines almost certainly was present in the antecedent primary tumors. Although larger confirmatory trials will be necessary, our findings suggest that epigenetic inactivation of hMLH1 underlies the pathogenesis of many human MSI colon cancers.
Colon cancer has served as a useful paradigm for understanding the molecular mechanisms of human carcinogenesis. In many colon cancers biallelic inactivation of two tumor suppressor genes, APC and p53, is demonstrable (1), and for these genes biallelic inactivation is usually caused by mutational inactivation of one allele, accompanied by deletion or loss of the cognate wild-type allele via mechanisms that cause loss of heterozygosity (1). In contrast, we found biallelic inactivation of the hMLH1 MMR gene commonly proceeds by epigenetic inactivation of both parental alleles and is not accompanied by loss of heterozygosity. These findings suggest that in these cancers epigenetic gene inactivation is a highly efficient method for silencing hMLH1 gene expression.
Previous studies of MSI colon cancers have demonstrated that these cancers as a class harbor increased methylation within promoter regions of multiple genes, including increased methylation of a candidate tumor suppressor gene, p16, as well as of a candidate tumor-promoting growth factor, the insulin-like growth factor II (20). It is thus possible that a large subset of MSI tumors harbors a general defect in gene regulation that induces aberrant promoter methylation at many loci. In this instance methylation and silencing of the hMLH1 promoter would provide a direct route that connects a general defect in promoter regulation with the specific MSI-associated pathway of carcinogenesis. This would be similar to the finding that mutational inactivation of a tumor suppressor gene, the type II transforming growth factor-β receptor, links the global mutator phenotype accompanying MSI with a defined pathway of malignant transformation (9–11). Future studies should reveal the specific relationship between epigenetic inactivation of the hMLH1 promoter and increased promoter methylation seen at other loci in some MSI cancers.
In addition to showing increased methylation of endogenous promoters, MSI cancers also have an increased propensity to methylate and silence exogenous retroviral gene sequences (21). This phenotype has most clearly been demonstrated for the HCT116 MSI colon cancer in which the hMLH1 gene is inactivated by mutation and in which the hMLH1 gene promoter is unmethylated. Future studies should elucidate whether the propensity to methylate exogenous gene sequences is or is not a reflection of the same disordered mechanism that underlies methylation and gene silencing of the hMLH1 locus. Additionally, increased methylation of the estrogen receptor promoter in normal colonic tissue has been reported to be a correlate of aging (22). Again, elucidating the relationship between this form of aberrant methylation and the epigenetic inactivation of the hMLH1 gene in cancer will be an important question for future investigation.
The causal role of MMR gene inactivation in initiating the pathway of MSI colon carcinogenesis suggests that in the tumors we studied the methylation and epigenetic silencing of the hMLH1 gene were neither consequences of disordered gene regulation in cancer nor results of the establishment of cancer cell lines. Instead, the epigenetic silencing of hMLH1 was a key pathophysiologic step in the genesis of these cancers. These findings support the idea that a similarly direct role in carcinogenesis is played by other tumor suppressor genes, such as p16 and VHL, in cancers in which those genes have been noted to be methylated and to additionally be inducible by 5-azacytidine (23–26).
Methylation of the hMLH1 gene promoter in association with absent hMLH1 protein was previously noted in several MSI colon cancers and in the AN3CA endometrial cancer (14). This current study establishes that hMLH1 promoter region methylation is strongly associated with epigenetic gene silencing, that the silencing mechanism is sufficient to accomplish biallelic inactivation of the hMLH1 gene, that silencing of the hMLH1 gene is reversible, that silencing of the hMLH1 gene appears to be actively maintained, and that hMLH1 silencing is a common route for the genesis of sporadic MSI cancers. Additionally, while this study was in progress, Herman et al. (27) also noted a significant incidence of hMLH1 promoter methylation among sporadic MSI colon cancers, as well as the ability to induce hMLH1 expression with demethylating agents. The large number of tumors analyzed in these three studies provides strong and consistent support for the contention that epigenetic silencing of hMLH1 is a common route leading to the genesis of sporadic MSI colon cancers. Also in support of this contention is the recent report that hMLH1 protein expression is absent by immunohistochemical assay in 90% of unselected human MSI colon cancers (28).
A singular finding of this current study is the observation that epigenetic inactivation of hMLH1 is biallelic and that reactivation of the hMLH1 allele is rapidly followed by resilencing of the gene. These observations suggest that hMLH1 silencing is the result of an underlying active mechanism that efficiently turns off hMLH1 gene expression. Moreover, it is highly tempting to suggest that hMLH1 promoter methylation is the direct mechanism that inactivates hMLH1 gene expression. However, both hypotheses require further investigation, and final judgment will have to await the ultimate elucidation of the mechanisms mediating hMLH1 promoter methylation and hMLH1 gene silencing.
Current data do not allow us to judge whether reexpression of wild-type hMLH1 will have therapeutic utility in certain MSI colon cancers, e.g., by reversing the resistance of such tumors to treatment with certain alkylating agents (29). Any trial of such therapy will need to reflect our finding that repetitive dosing of 5-azacytidine is required to maintain hMLH1 expression. It is also tempting to consider that demethylating agents might play a role in cancer prevention in individuals who are at risk for cancer or in individuals in whom hMLH1 promoter methylation might be detected as an early neoplastic change. Our data support the suggestion that hMLH1 gene silencing accounts for a significant number of human colon cancers. Additionally, the epigenetic inactivation of hMLH1 demonstrated in the AN3CA MSI endometrial cancer may prove to be representative of MSI endometrial cancers, which as a class bear mostly wild-type MMR alleles (13). Finally, many sporadic human gastric cancers also arise via the MSI pathway (2). Elucidating the ultimate mechanism underlying the epigenetic inactivation of hMLH1 expression may thus prove relevant to the understanding of many different types of human cancer.
Acknowledgments
We thank James K. V. Willson for establishing the Vaco cell lines and Bert Vogelstein for his help in determining the MSI status and the sequences of the MMR genes in the Vaco cell lines and also for many helpful discussions. We also thank James Herman and Bo Liu for helpful discussions and Jen Sedwick for help with manuscript preparation. This work was supported by Grant CA 67409 (to S.D.M.), Grant CA 72160 (to S.D.M.), Grant CA 70788 (to M.L.V.), and Grant CA 4370301 (to the Case Western Reserve University Cancer Center) from the National Institutes of Health, Grant FRA-451 (to S.D.M.) from American Cancer Society, Grant 96B084 (to W.D.S.) from American Institute for Cancer Research, and from funds from the State of Ohio Board of Regents. S.D.M. and P.M. are Investigators of the Howard Hughes Medical Institute.
ABBREVIATIONS
- MMR
mismatch repair
- MSI
microsatellite instability
References
- 1.Kinzler K, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Eshleman J, Markowitz S. Curr Opin Oncol. 1995;7:83–89. [PubMed] [Google Scholar]
- 3.Marra G, Boland C R. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 4.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 5.Perucho M. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 6.Leach F, Nicolaides N, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen L, Nyström-Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 7.Parsons R, Li G-M, Longley M J, Fang W H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 8.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 9.Eshleman J, Lang E, Bowerfind G, Parsons R, Vogelstein B, Willson J, Veigl M, Sedwick W D, Markowitz S. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 10.Bhattacharyya N, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R, Zborowska E, Kinzler K, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 12.Liu B, Nicolaides N, Markowitz S, Willson J, Parsons R, Jen J, de la Chapelle A, Hamilton S, Kinzler K, Vogelstein B. Nat Genet. 1995;9:48–53. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 13.Katabuchi H, van Rees B, Lambers A R, Ronnett B M, Blazes M S, Leach F S, Cho K R, Hedrick L. Cancer Res. 1995;55:5556–5560. [PubMed] [Google Scholar]
- 14.Kane M, Loda M, Gaida G, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 15.Willson J, Bittner G, Oberley T, Meisner G, Weese J. Cancer Res. 1987;47:2704–2713. [PubMed] [Google Scholar]
- 16.Davis L, Dibner M, Battey J. Basic Methods in Molecular Biology. New York: Elsevier Science; 1986. p. 388. [Google Scholar]
- 17.Boyer J C, Umar A, Risinger J I, Lipford J R, Kane M, Yin S, Barrett J C, Kolodner R D, Kunkel T A. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 18.Drummond J, Anthony A, Brown R, Modrich P. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 19.Li G-M, Modrich P. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahuja N, Mohan A, Li Q, Stolker J, Herman J, Hamilton S, Baylin S, Issa J-P. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 21.Lengauer C, Kinzler K, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa J-P, Ottaviano Y, Celano P, Hamilton S, Davidson N, Baylin S. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 23.Herman J, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M, et al. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 25.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Zuleta M, Bender C, Yang A, Nguyen T, Beart R, Van Tornout J, Jones P. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 27.Herman J G, Umar A, Polyak K, Graff J, Ahuja N, Issa J-P, Markowitz S, Willson J K V, Hamilton S, Kinzler K, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thibodeau S, French A, Cunningham J, Tester D, Burgart L, Roche P, McDonnell S, Schaird D, Vockley C, Michels V, et al. Cancer Res. 1998;58:1713–1718. [PubMed] [Google Scholar]
- 29.Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]