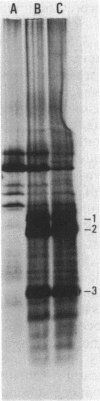
Abstract
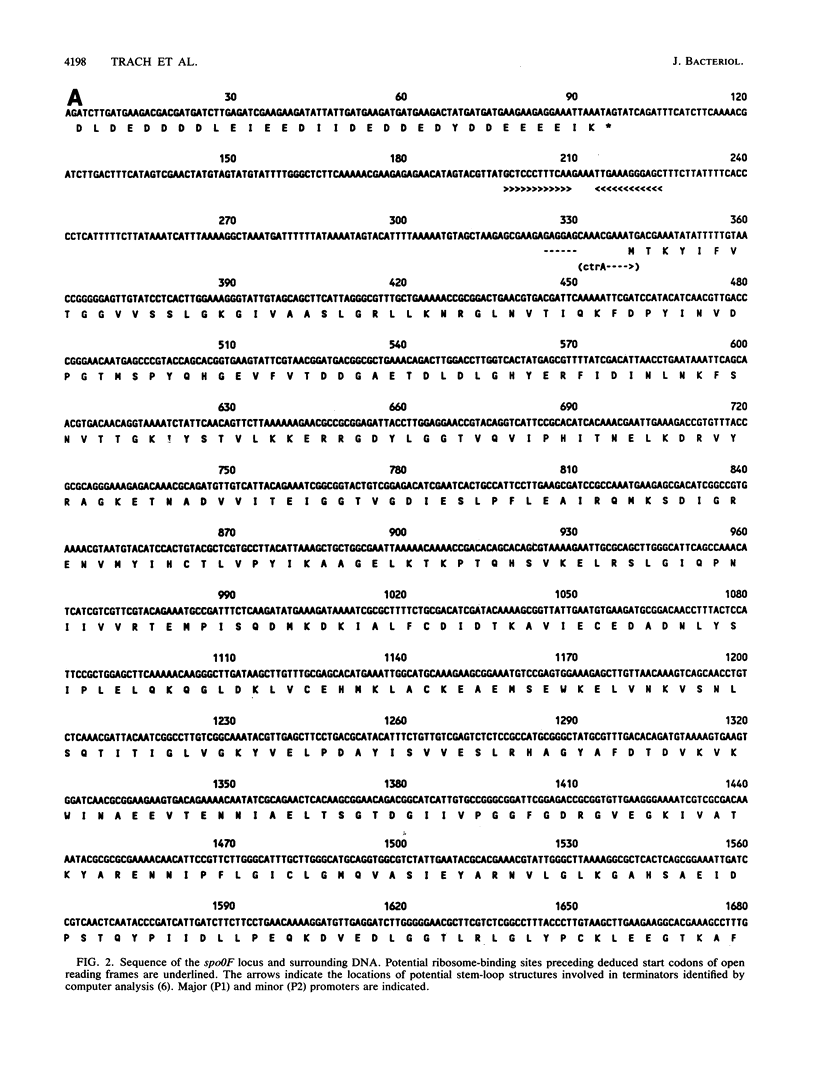
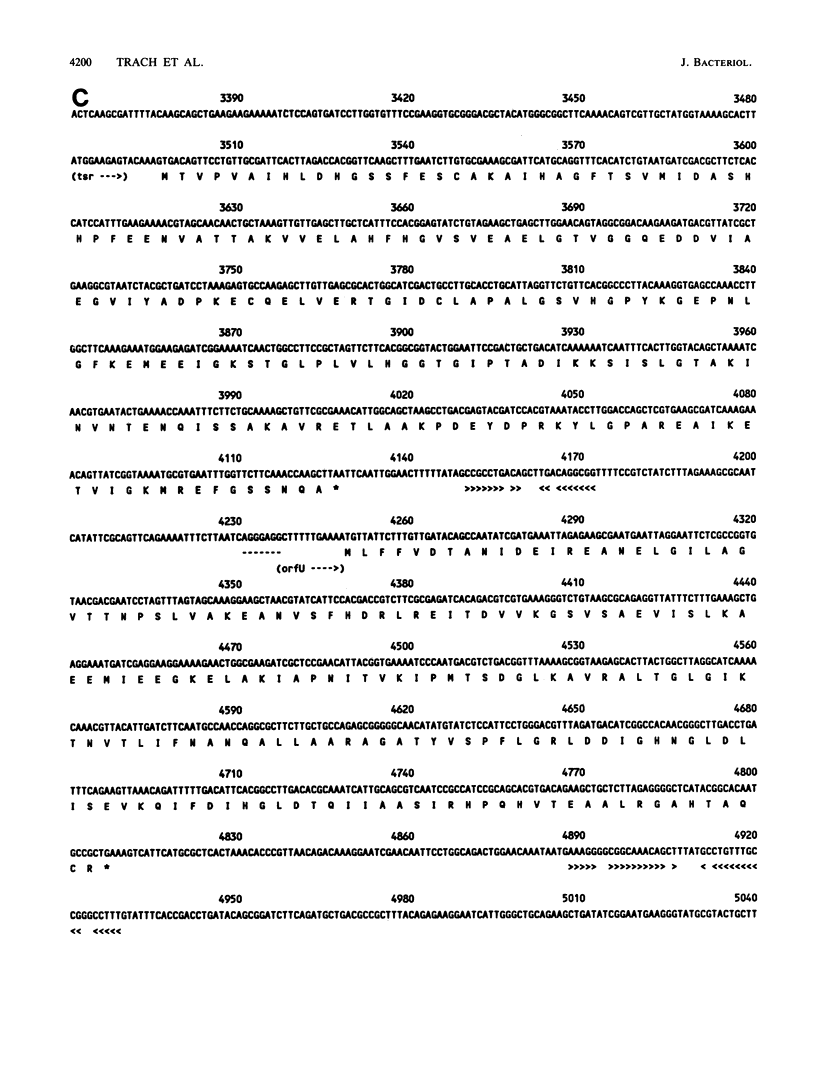
The total sequence of a 6,314-base-pair BglII fragment of the Bacillus subtilis chromosome containing the spo0F locus has been accomplished. Several genes of interest have been identified on this DNA fragment. The ctrA locus was recognized as coding for CTP synthetase by comparison of its deduced sequence with that of Escherichia coli CTP synthetase. A total of 53% of the residues are identical between the enzymes from these organisms. The spo0F locus was followed immediately by a locus, tsr, required for RNA synthesis in this organism. Temperature-sensitive mutations within the tsr locus have been identified, but strains with deletions of the locus are nonviable. It was concluded that tsr codes for an unknown essential component of the RNA synthesis machinery. The tsr gene was followed by another open reading frame which could code for a protein of 19,975 Mr. This gene was translated in vivo, but deletion-insertion mutations within the gene had no phenotype. The gene was cotranscribed with the tsr gene, although about 50% of the transcripts terminated between the two genes. The rev-4 mutation which reverts the sporulation-defective phenotype of erythromycin-resistant mutants was located to a partial open reading frame at the end of the fragment. Disruption of this open reading frame by deletion-insertion mutation did not result in a discernible phenotype. S1 protection experiments located the start sites of transcription for several of the genes on this fragment. The spo0F gene was found to be monocistronic. Regulation of the identified genes was investigated by using beta-galactosidase gene fusions.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier J., Stragier P., Bonamy C., Szulmajster J. Nucleotide sequence of the spo0B gene of Bacillus subtilis and regulation of its expression. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7012–7016. doi: 10.1073/pnas.81.22.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm S. P., Staal S. P., Hoch J. A. Phenotypes of pleiotropic-negative sporulation mutants of Bacillus subtilis. J Bacteriol. 1973 Sep;115(3):1063–1070. doi: 10.1128/jb.115.3.1063-1070.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., Hoch J. A. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J Bacteriol. 1985 Feb;161(2):556–562. doi: 10.1128/jb.161.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Hoch J. A., Trach K., Kawamura F., Saito H. Identification of the transcriptional suppressor sof-1 as an alteration in the spo0A protein. J Bacteriol. 1985 Feb;161(2):552–555. doi: 10.1128/jb.161.2.552-555.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh J., Ikeuchi T., Kurahashi K. Nucleotide sequences of the sporulation gene spo0A and its mutant genes of Bacillus subtilis. Proc Natl Acad Sci U S A. 1985 May;82(9):2665–2668. doi: 10.1073/pnas.82.9.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
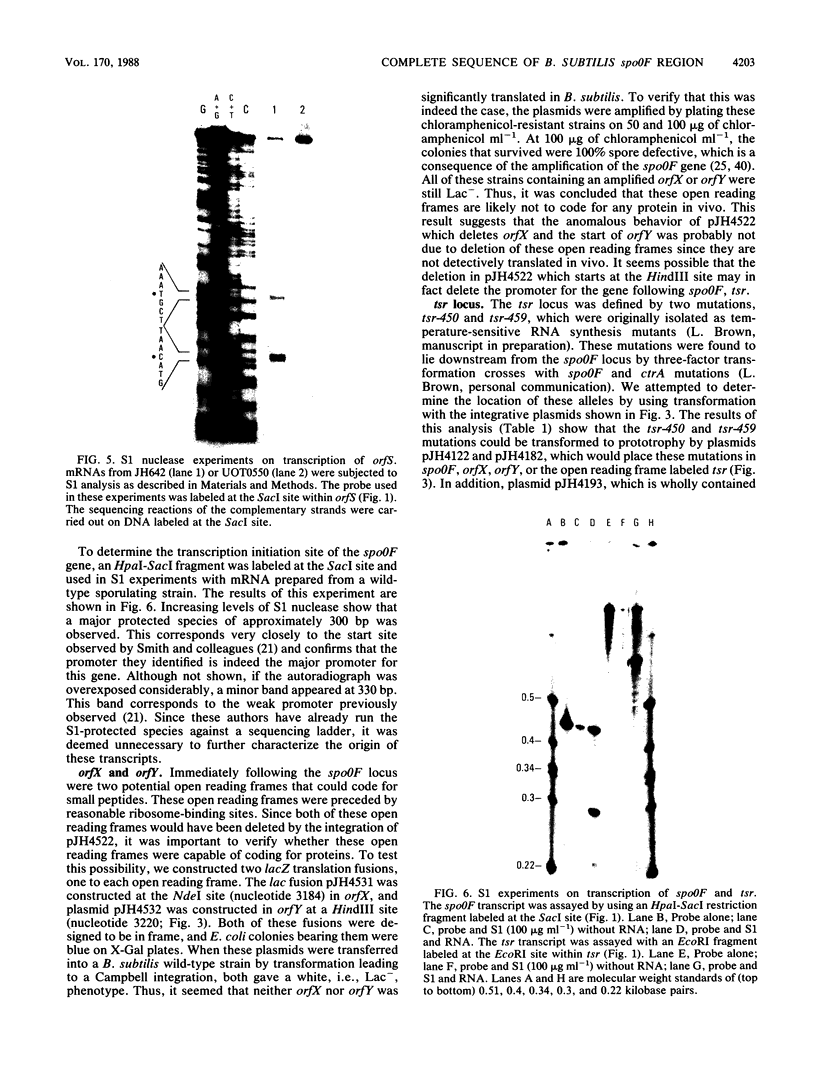
- Lewandoski M., Dubnau E., Smith I. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J Bacteriol. 1986 Nov;168(2):870–877. doi: 10.1128/jb.168.2.870-877.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Kawamura F., Kobayashi Y., Saito H. Early sporulation gene spo0F: nucleotide sequence and analysis of gene product. Proc Natl Acad Sci U S A. 1983 Feb;80(3):658–662. doi: 10.1073/pnas.80.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stone A. B. A simplified method for preparing sucrose gradients. Biochem J. 1974 Jan;137(1):117–118. doi: 10.1042/bj1370117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Makaroff C. A., Zalkin H. Nucleotide sequence of Escherichia coli pyrG encoding CTP synthetase. J Biol Chem. 1986 Apr 25;261(12):5568–5574. [PubMed] [Google Scholar]
- Yamashita S., Yoshikawa H., Kawamura F., Takahashi H., Yamamoto T., Kobayashi Y., Saito H. The effect of spo0 mutations on the expression of spo0A- and spo0F-lacZ fusions. Mol Gen Genet. 1986 Oct;205(1):28–33. doi: 10.1007/BF02428029. [DOI] [PubMed] [Google Scholar]
- Yang M., Ferrari E., Chen E., Henner D. J. Identification of the pleiotropic sacQ gene of Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):113–119. doi: 10.1128/jb.166.1.113-119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Kazami J., Yamashita S., Chibazakura T., Sone H., Kawamura F., Oda M., Isaka M., Kobayashi Y., Saito H. Revised assignment for the Bacillus subtilis spo0F gene and its homology with spo0A and with two Escherichia coli genes. Nucleic Acids Res. 1986 Jan 24;14(2):1063–1072. doi: 10.1093/nar/14.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]