Abstract
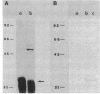
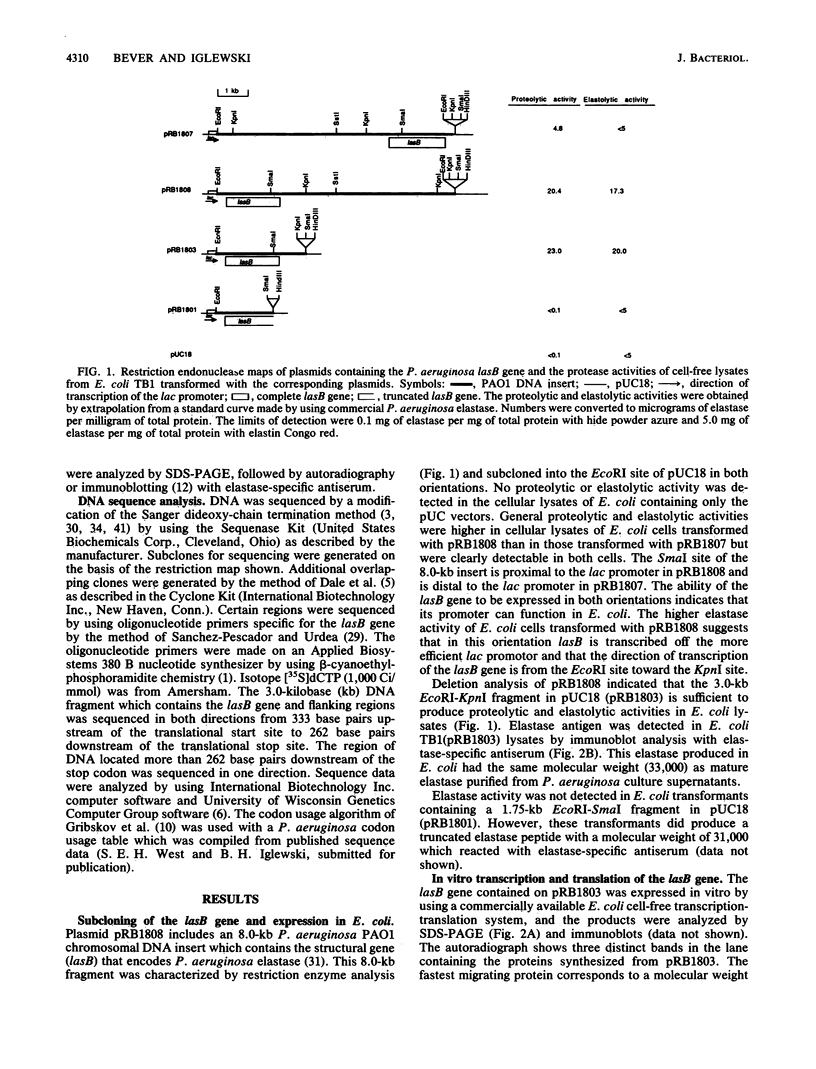
The elastase structural gene (lasB) from Pseudomonas aeruginosa PAO1 has been previously cloned on an 8-kilobase (kb) DNA fragment. The lasB gene, cloned in both orientations in pUC18, produced elastase in Escherichia coli, indicating that its promoter and translation initiation sites are functional in E. coli. Deletion analysis further defined the location of the lasB gene to a 3.0-kb EcoRI-KpnI fragment (pRB1803). Elastase prepared from E. coli TB1 (pRB1803) corresponded in molecular weight to mature P. aeruginosa extracellular elastase (33,000). The lasB gene directed the synthesis of 54- and 50-kilodalton (kDa) proteins in a bacterial cell-free transcription-translation system. The 33-, 50-, and 54-kDa proteins reacted with elastase-specific antiserum. To further characterize the lasB gene, the nucleotide sequence of the 3.0-kb EcoRI-KpnI fragment was determined. This DNA fragment contained a 1,491-base-pair open reading frame encoding 498 amino acids, corresponding to a predicted molecular weight of 53,600. The deduced amino acid sequence contained a putative signal sequence followed by a large polypeptide which preceded the mature 33-kDa elastase protein. Three zinc ligands and an active site were predicted for the mature elastase on the basis of its amino acid sequence homology with Bacillus thermoproteolyticus thermolysin.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring G., Obernesser H. J., Botzenhart K. Extrazelluläre Toxine von Pseudomonas aeruginosa. II. Einwirkung zweier gereinigter Proteasen auf die menschlichen Immunoglobuline IgG, IgA und sekretorisches IgA. Zentralbl Bakteriol A. 1981 Mar;249(1):89–98. [PubMed] [Google Scholar]
- Fecycz I. T., Campbell J. N. Mechanisms of activation and secretion of a cell-associated precursor of an exocellular protease of Pseudomonas aeruginosa 34362A. Eur J Biochem. 1985 Jan 2;146(1):35–42. doi: 10.1111/j.1432-1033.1985.tb08616.x. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Activation of an elastase precursor by the lasA gene product of Pseudomonas aeruginosa. J Bacteriol. 1987 Oct;169(10):4532–4539. doi: 10.1128/jb.169.10.4532-4539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck L. W., Morihara K., McRae W. B., Miller E. J. Specific cleavage of human type III and IV collagens by Pseudomonas aeruginosa elastase. Infect Immun. 1986 Jan;51(1):115–118. doi: 10.1128/iai.51.1.115-118.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindahl M. S., Iglewski B. H. Outer membrane proteins from Legionella pneumophila serogroups and other Legionella species. Infect Immun. 1986 Jan;51(1):94–101. doi: 10.1128/iai.51.1.94-101.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes M. A., Matthews B. W. Structure of thermolysin refined at 1.6 A resolution. J Mol Biol. 1982 Oct 5;160(4):623–639. doi: 10.1016/0022-2836(82)90319-9. [DOI] [PubMed] [Google Scholar]
- Jensen S. E., Phillippe L., Teng Tseng J., Stemke G. W., Campbell J. N. Purification and characterization of exocellular proteases produced by a clinical isolate and a laboratory strain of Pseudomonas aeruginosa. Can J Microbiol. 1980 Jan;26(1):77–86. doi: 10.1139/m80-012. [DOI] [PubMed] [Google Scholar]
- Kessler E., Safrin M. Partial purification and characterization of an inactive precursor of Pseudomonas aeruginosa elastase. J Bacteriol. 1988 Mar;170(3):1215–1219. doi: 10.1128/jb.170.3.1215-1219.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu P. V. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. II. Effects of lecithinase and protease. J Infect Dis. 1966 Feb;116(1):112–116. doi: 10.1093/infdis/116.1.112. [DOI] [PubMed] [Google Scholar]
- MORIHARA K. PRODUCTION OF ELASTASE AND PROTEINASE BY PSEUDOMONAS AERUGINOSA. J Bacteriol. 1964 Sep;88:745–757. doi: 10.1128/jb.88.3.745-757.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W., Weaver L. H., Kester W. R. The conformation of thermolysin. J Biol Chem. 1974 Dec 25;249(24):8030–8044. [PubMed] [Google Scholar]
- Morihara K., Tsuzuki H., Harada M., Iwata T. Purification of human plasma alpha 1-proteinase inhibitor and its inactivation by Pseudomonas aeruginosa elastase. J Biochem. 1984 Mar;95(3):795–804. doi: 10.1093/oxfordjournals.jbchem.a134671. [DOI] [PubMed] [Google Scholar]
- Morihara K., Tsuzuki H., Oda K. Protease and elastase of Pseudomonas aeruginosa: inactivation of human plasma alpha 1-proteinase inhibitor. Infect Immun. 1979 Apr;24(1):188–193. doi: 10.1128/iai.24.1.188-193.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahama K., Yoshimura K., Marumoto R., Kikuchi M., Lee I. S., Hase T., Matsubara H. Cloning and sequencing of Serratia protease gene. Nucleic Acids Res. 1986 Jul 25;14(14):5843–5855. doi: 10.1093/nar/14.14.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Cryz S. J., Iglewski B. H. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980 Jun;142(3):836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pescador R., Urdea M. S. Use of unpurified synthetic deoxynucleotide primers for rapid dideoxynucleotide chain termination sequencing. DNA. 1984 Aug;3(4):339–343. doi: 10.1089/dna.1.1984.3.339. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad P. A., Bever R. A., Nicas T. I., Leduc F., Hanne L. F., Iglewski B. H. Cloning and characterization of elastase genes from Pseudomonas aeruginosa. J Bacteriol. 1987 Jun;169(6):2691–2696. doi: 10.1128/jb.169.6.2691-2696.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad P. A., Iglewski B. H. Nucleotide sequence and expression in Escherichia coli of the Pseudomonas aeruginosa lasA gene. J Bacteriol. 1988 Jun;170(6):2784–2789. doi: 10.1128/jb.170.6.2784-2789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. R., Miller K. D. Elastase of Pseudomonas aeruginosa: inactivation of complement components and complement-derived chemotactic and phagocytic factors. Infect Immun. 1974 Jul;10(1):128–135. doi: 10.1128/iai.10.1.128-135.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. H., Kester W. R., Matthews B. W. A crystallographic study of the complex of phosphoramidon with thermolysin. A model for the presumed catalytic transition state and for the binding of extended substances. J Mol Biol. 1977 Jul;114(1):119–132. doi: 10.1016/0022-2836(77)90286-8. [DOI] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. E., Iglewski B. H. Toxins of Pseudomonas aeruginosa: new perspectives. Rev Infect Dis. 1983 Sep-Oct;5 (Suppl 4):S715–S722. doi: 10.1093/clinids/5.supplement_4.s715. [DOI] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]