Abstract
Bloom’s syndrome (BS) and Werner’s syndrome (WS) are genetic disorders in which an increased rate of chromosomal aberration is detected. The genes responsible for these diseases, BLM and WRN, have been found to be homologs of Escherichia coli recQ and Saccharomyces cerevisiae SGS1 genes. Here we show that yeast Sgs1 helicase acts as a suppressor of illegitimate recombination through homologous recombination and that human BLM and WRN helicases can suppress the increased homologous and illegitimate recombinations in the S. cerevisiae sgs1 mutant. The results imply a role of BLM and WRN helicases to control genomic stability in human cells. Similar to Sgs1 helicase, BLM helicase suppressed the cell growth in the top3 sgs1 mutation background and restored the increased sensitivity of the sgs1 mutant to hydroxyurea, but the WRN helicase did not. We discussed differential roles of BLM and WRN helicases in human cells. BLM- and WRN-bearing yeasts provide new useful models to investigate human BS and WS diseases.
Keywords: RecQ homologue/Rqh1/illegitimate recombination/homologous recombination/topoisomerase III
Bloom’s syndrome (BS) is an autosomal recessive disorder causing short stature, immunodeficiency, and an increasing risk of cancer. BS cells have a high level of genomic instability at the cellular level, as shown by an increased rate of sister chromatid exchange and chromosomal aberration (1). Werner’s syndrome (WS) also is a rare genetic disorder and is characterized by the premature appearance of normal aging in young adults. Cultured cells derived from patients with WS show an increased rate of somatic mutations, chromosome losses, and deletions (2–4). These experiments may suggest an increased rate of illegitimate recombination in both BS and WS cells. Recently, the genes BLM and WRN, responsible for BS and WS, respectively, have been found to be homologs of Escherichia coli recQ and Saccharomyces cerevisiae SGS1 genes, which encode DNA helicases (5–9). In E. coli, RecQ helicase is involved in homologous and illegitimate recombination (7, 10). In yeast, Sgs1 protein physically interacts with topoisomerase II and III (11, 12). The mutation of the SGS1 gene suppresses the growth defect generated by top3 mutation and also shows an increased recombination at the repeated ribosomal DNA locus and other loci (9, 12). The sgs1 mutation also reduces the average lifespan of yeast cells while it enhances relocalization of Sir proteins to nucleolus and accumulation of extrachromosomal rDNA circles (13, 14). But whether the sgs1 mutation affects illegitimate recombination is unknown.
Illegitimate recombination takes place between nonhomologous DNA sequences or very short regions of homology. Nonhomologous end-joining, which is known to be mediated by Rad50, Xrs2, Mre11, Hdf1, Ku80, and Dnl4 as well as silencing factors Sir2, Sir3, and Sir4 (15–24), has an essential role in illegitimate recombination. In this paper, we show that yeast Sgs1 helicase acts as a suppressor of illegitimate recombination through homologous recombination and that human BLM and WRN helicases can suppress the increased homologous and illegitimate recombination in the yeast sgs1 mutant. In addition, BLM helicase suppressed the cell growth in the top3 sgs1 background and restored the increased sensitivity of the sgs1 mutant to hydroxyurea, but the WRN helicase did not.
MATERIALS AND METHODS
Strains and Plasmids.
Yeast strains DH6.61D (MATa trp1 his3 leu2 ura3 can1 cyh2) and its derivatives, constructed by one-step gene replacement (25), were used for biological assays described below. The SGS1, TOP3, and RAD52 genes were replaced by sgs1∷TRP1, top3∷LEU2, and rad52∷HIS3, respectively. YT444 hdf1∷LEU2 was described by Tsukamoto et al. (16). KY21 sgs1∷BLM+ and KY22 sgs1∷BLM− were constructed by replacement of the SGS1 gene with sgs1∷GAPDH promoter-BLM cDNA (same or opposite direction to the promoter)-ADH terminator-TRP1, respectively. KY23 sgs1∷WRN+ and KY24 sgs1∷WRN− were constructed in a similar way to KY21 and KY22. YCpL2 was described in Tsukamoto et al. (15). YCpHR was constructed as follows. A XhoI fragment containing the CAN1, LEU2, URA3, and TRP1 genes of YCpL2 (15) was ligated with a SalI-XhoI fragment containing the LEU2 gene of YEp13 plasmid (26). The resulting plasmid YCpHR contains two LEU2 genes with the same orientation.
A WRN cDNA fragment containing the complete ORF was cloned by reverse transcription–PCR, using HeLa mRNA as a template, and constructed as follows. Based on the nucleotide sequence of the WRN cDNA (6), a sense primer, 5′-TGAAGACATTGTTTTTTGGACTCTGC-3′, and an antisense primer, 5′-GGATAGATTAAGTTTCCTAAGTTCACC-3′, were used to amplify 5′ half of the cDNA. A sense primer, 5′-CCATTTCTTGTCAAAACAAGTTCCC-3′, and an antisense primer, 5′-CATAATTGTTCTGGTAATTGCCAGC-3′, were used to amplify 3′ half of the cDNA. The WRN cDNA containing the complete ORF was constructed by ligation at XbaI sites contained in an overlapped region of both 5′ and 3′ half fragments. Similarly, based on the nucleotide sequence of the BLM cDNA (5), 5′ half fragment of BLM cDNA was cloned by using a sense primer, 5′-ATAGTCTGCGTGCGAGGATTATGGC-3′ and an antisense primer, 5′-CATCTCTCTGTAACGTGTCAGCC-3′ for amplification. 3′ half fragment of BLM cDNA was cloned by using a sense primer, 5′-TCTCCAGGCGAGAATGTGACACC-3′, and an antisense primer, 5′-GACAAACAAGAAAGAGGGTCTATG-3′, for amplification. The BLM cDNA containing the complete ORF was constructed by ligation at NcoI sites contained in an overlapped region of both 5′ and 3′ half fragments. The nucleotide sequences of the resulting cDNAs determined by the standard method indicated that the cDNAs do not have any mutation.
Recombination Assays.
Determinations of the rate of illegitimate recombination in plasmid YCpL2 and detection of plasmid deletion by end-joining using dicentric plasmid YdCp2 were made as described by Tsukamoto et al. (15, 16). Determinations of the rates of homologous recombination between direct repeats in plasmid YCpHR were made by fluctuation analysis (27).
Measurement of Growth Rate.
DH6.61D and its derivatives were streaked onto a yeast extract/peptone/dextrose agar plate and were photographed after growth for 2 days at 30°C.
Measurement of Sensitivity to Hydroxyurea.
Exponential cultures of DH6.61D and its derivatives were diluted serially by 10-fold and were spotted onto synthetic complete plates with or without hydroxyurea (100 mM). The plates were then incubated at 30°C for 2 days.
Determination of Sequences of Recombination Junctions.
The location of recombination junctions in YCpL2 plasmid was determined by using sets of primers as described in Tsukamoto et al. (15). A DNA sequence containing a recombination junction was amplified by using the PCR with Taq polymerase. The amplified DNA fragment was cloned into pUC119 plasmid, and the nucleotide sequence was determined by the dideoxy termination method using A.L.F. DNA sequencer (Pharmacia).
Western Blot Analysis.
Total cell extracts were prepared by disrupting cells with glass beads. The samples were then electrophoresed in a 7.5% SDS-polyacrylamide gel, transferred to poly(vinylidene difluoride) membranes, and probed with 1:2 diluted anti-WRN helicase mouse IgG. As a secondary antibody, alkaline phosphatase-conjugated goat anti-mouse IgG antibody (Kirkegaard & Perry Laboratories) was used. Anti-WRN helicase IgG used as a primary antibody was a mouse monoclonal IgG and was prepared by using a recombinant C-terminal fragment of WRN helicase expressed in E. coli.
RESULTS
Yeast sgs1 Mutation Enhances Illegitimate Recombination.
To study the role of Sgs1 helicase in illegitimate recombination, we examined the effect of the sgs1 mutation on generating deletions by using a YCp plasmid, YCpL2 (15). The YCpL2 plasmid carries two negative selection markers, the CAN1 and CYH2 genes. A can1 cyh2 strain containing YCpL2 shows sensitivity to both canavanine and cycloheximide, and deletions generated in the CAN1 and CYH2 genes on YCpL2 can be detected by selecting Ura+ colonies resistant to both drugs. The plasmids isolated by this selection have been shown previously to be formed by illegitimate recombination (15). The YCpL2 was introduced into a sgs1 mutant, and the rate of illegitimate recombination was determined. The rate in the sgs1 mutant was 11-fold higher than that in the wild-type strain (Table 1), which shows that Sgs1 helicase has a role as a suppressor of illegitimate recombination.
Table 1.
Effect of sgsI mutation on illegitimate recombination in YCpL2 plasmid
| Strain | Genotype | Recombination rate, 10−7, CanR CyhR/cell per generation | Fold increase |
|---|---|---|---|
| DH6.61D | Wild type | 1.6 (0.18) | 1 |
| KY11 | rad52 | 0.051 (0.039) | 0.031 |
| YT444 | hdf1 | 0.033 (0.012) | 0.021 |
| KY12 | sgs1 | 17 (0.79) | 11 |
| KY13 | sgs1 rad52 | 0.43 (0.16) | 0.27 |
| KY14 | sgs1 hdf1 | 0.043 (0.019) | 0.027 |
Recombination rates were determined by fluctuation analysis of deletion formation in YCpL2 plasmid as described in Tsukamoto et al. (15). SEM is shown in parentheses.
The recombinant plasmids obtained from five CanR CyhR cells of the sgs1 mutant were rescued. The recombination junctions were mapped by PCR, and the nucleotide sequences at the recombination junctions were determined. The nucleotide sequences of the parental recombination sites also were determined. As shown in Fig. 1, there are short regions of homology at most recombination sites. With respect to one plasmid, SR1, there is no homologous region at the junction of that plasmid. These results confirmed that the deletions are caused by illegitimate recombination.
Figure 1.
Junction sequences of deleted plasmids derived from the sgs1 mutant. The top sequence represents the parental sequence corresponding to the left side of the junction, the middle sequence represents the sequence of the recombinant, and the bottom sequence represents the parental sequence corresponding to the right side of the junction. Homologous sequences around a junction are represented by a bold letter. The orientation of DNA is 5′ to 3′ from left to right. Numbers represent the map coordinates of the YCpL2 sequence.
The Increase of Illegitimate Recombination in the sgs1 Mutant Takes Place Through Homologous Recombination.
Recently, Tsukamoto et al. (15, 16) indicated that Rad52 and Hdf1, which are involved in homologous recombination (28) and end-joining (16, 17), respectively, participate in deletions of the YCpL2 plasmid (see also Table 1). This controversial result is explained by a model, in which unstable dicentric plasmids formed by the Rad52-dependent homologous recombination between two replicated plasmids could be broken subsequently between two centromeres, resulting in deletions by the Hdf1-dependent end-joining. To study whether the illegitimate recombination enhanced by the sgs1 mutation also stems from homologous recombination and end-joining, we examined the requirement of Rad52 and Hdf1 in the deletion events enhanced by sgs1 mutation in the YCpL2 plasmid. The rates of recombination were reduced 40- and 400-fold in the sgs1 rad52 and sgs1 hdf1 double mutants, respectively, compared with that of the sgs1 single mutant (Table 1). These results indicate that the majority of illegitimate recombination enhanced by the sgs1 mutation depends on the Rad52 and Hdf1 functions and therefore is mediated by homologous recombination and end-joining processes, consistent with Tsukamoto’s model (18).
In addition, the frequency of illegitimate recombination in the sgs1 rad52 mutant also is increased 8.4-fold compared with that in the rad52 mutant, indicating that the Rad52-independent illegitimate recombination is increased by the sgs1 mutation, although the frequency of illegitimate recombination is about 30-fold lower than that in the RAD52+ cell (Table 1). It is not clear yet whether the minor illegitimate recombination pathway in the sgs1 rad52 genetic background takes place through Rad52-independent homologous recombination or whether this pathway takes place by illegitimate recombination not involving a homologous recombination process. Our further studies are concentrated on the major illegitimate recombination pathway that takes place through Rad52-dependent homologous recombination.
To determine whether Sgs1 helicase is involved in dimer formation by Rad52-dependent homologous recombination or end-joining, we examined the effect of sgs1 mutation on a deletion assay by using a dicentric plasmid, YdCp2 (18), in which double-strand breaks are expected to occur at a high frequency because the plasmid contains two centromeres and is structurally unstable. Therefore, this assay can be used as an indicator of the end-joining reaction. The frequency of deletion in the sgs1 mutant was comparable to that in the wild-type strain (data not shown). Therefore, this result suggests that Sgs1 helicase is not involved in end-joining, but in dimer formation, which is mediated by homologous recombination. This result is consistent with the notion that Sgs1 helicase participates in the suppression of homologous recombination (9, 12). From these results, we infer that Sgs1 helicase has a role as a suppressor of illegitimate recombination by regulating homologous recombination for dimer formation in the major illegitimate recombination pathway.
Human BLM or WRN Helicase Can Suppress the Increased Illegitimate and Homologous Recombinations in the Yeast sgs1 Mutant.
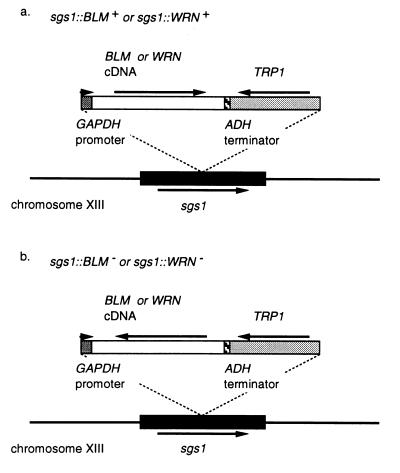
Because BS and WS cells show the phenotype of genomic instability, BLM and WRN genes may have a role in controlling illegitimate recombination in human cells. To examine whether human BLM or WRN helicase can suppress the hyperrecombination phenotype of the sgs1 mutant, we constructed two sets of the sgs1 disruptants, sgs1∷BLM+ and sgs1∷BLM−, and sgs1∷WRN+ and sgs1∷WRN−. In the sgs1∷BLM+ strains, the BLM cDNA was located downstream of GAPDH promoter in the correct direction, whereas in the sgs1∷BLM− strains, it was located in the opposite direction of the promoter (Fig. 2). The sgs1∷WRN+ and sgs1∷WRN− strains also were constructed in the same way as sgs1∷BLM+ and sgs1∷BLM−, respectively.
Figure 2.
Construction of BLM- and WRN-bearing yeast strains. (a) Structures of the sgs1∷BLM+ and sgs1∷WRN+strains. To construct these strains, we inserted a GAPDH promoter-BLM cDNA (or WRN cDNA)-ADH terminator-TRP1 fragment into the SGS1 locus of S. cerevisiae DH6.61D by a one-step gene replacement method (25). The BLM cDNA (or WRN cDNA) was connected with the GAPDH promoter in the correct orientation. Arrows indicate the orientation of GAPDH promoter and other genes. (b) Structures of the sgs1∷BLM− and sgs1∷WRN− strains. The sgs1∷BLM− and sgs1∷WRN− strains are also constructed by insertion into the SGS1 locus as shown above, except that the BLM cDNA (or WRN cDNA) was connected with the GAPDH promoter in the opposite orientation.
Because it has been observed that overexpression of Sgs1 in yeast is toxic and leads to fragmentation of the nucleus (12, 13), we examined the growth rate of these strains. The doubling time of the wild type, sgs1, sgs1∷BLM+, sgs1∷BLM−, sgs1∷WRN+, or sgs1∷WRN− was 105, 108, 94, 92, 102, or 99 min, respectively. The growth rates of the sgs1∷BLM+ and sgs1∷BLM− strains and those of the sgs1∷WRN+ and sgs1∷WRN− strains were comparable with each other, indicating that expression of WRN or BLM helicase in yeast does not affect the cell growth.
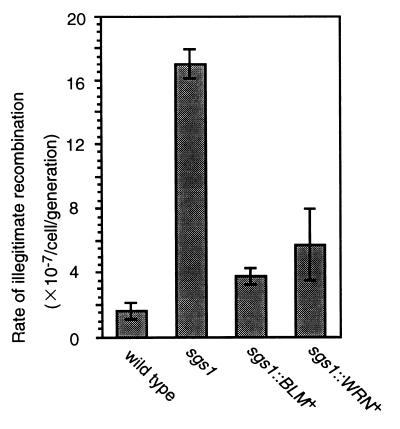
Then, we examined the rate of illegitimate recombination in the sgs1∷BLM+ and sgs1∷WRN+ strains by using YCpL2 plasmid. The rate of illegitimate recombination in the sgs1∷BLM+ strain was 4.5-fold lower than that in the sgs1 mutant and was 2.4-fold higher than that in the wild-type strain (Fig. 3). A similar result was obtained with the sgs1∷WRN+ strain (Fig. 3). These results indicate that the human BLM and WRN helicases can function as suppressors of illegitimate recombination, although the compensation of the sgs1 mutation was partial.
Figure 3.
The rate of illegitimate recombination in the sgs1∷BLM+ and sgs1∷WRN+strains. The rates of illegitimate recombination in DH6.61D wild type, KY12 sgs1, KY21 sgs1∷BLM+, and KY23 sgs1∷WRN+ were determined by fluctuation analysis by using plasmid YCpL2 as described in Table 1. Vertical bars represent SEM.
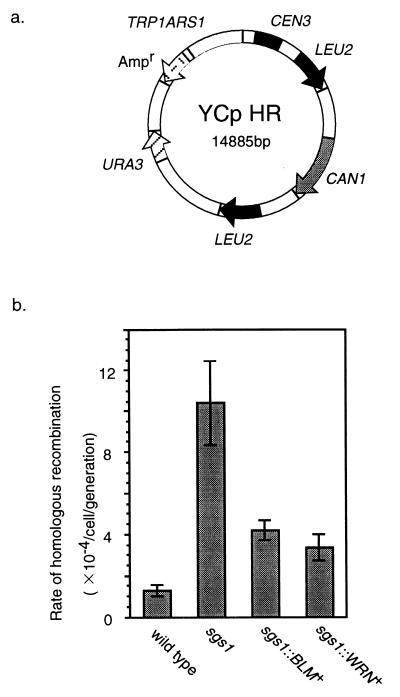
To examine whether BLM and WRN helicases can suppress homologous recombination in the sgs1 mutant, we constructed a plasmid, YCpHR, to detect homologous recombination between direct repeats (Fig. 4a). The YCpHR carries a negative selection marker, the CAN1 gene. A can1 mutant containing YCpHR shows sensitivity to canavanine, and deletions generated by homologous recombination between the LEU2 repeats can be monitored by counting the drug-resistant Ura+ colonies. The YCpHR was introduced into the sgs1 mutant, and the rates of homologous recombination were determined. We confirmed that the sgs1 mutant shows the hyperrecombination phenotype by an assay with YCpHR (Fig. 4b). Next, we examined the rate of homologous recombination in the sgs1∷BLM+ and sgs1∷WRN+ strains. The rate of recombination in the sgs1∷BLM+ strain was 2.4-fold lower than that in the sgs1 mutant and was 3.2-fold higher than that in the wild-type strain (Fig. 4b). A similar result was obtained with the sgs1∷WRN+ strain (Fig. 4b). These results show that human BLM and WRN helicases can function as suppressors of homologous recombination, although the BLM and WRN genes cannot completely substitute for the defect in the sgs1 strain.
Figure 4.
The rate of homologous recombination in the sgs1∷BLM+ and sgs1∷WRN+ strains. (a) Plasmid YCpHR carries the CAN1 gene between direct repeats of the LEU2 gene and is used to determine the rate of homologous recombination. (b) The rates of deletion between direct repeats in plasmid YCpHR. The rates of homologous recombination between direct repeats in DH6.61D wild type, KY12 sgs1, KY21 sgs1∷BLM+, and KY23 sgs1∷WRN+ were determined as described above. Vertical bars represent SEM.
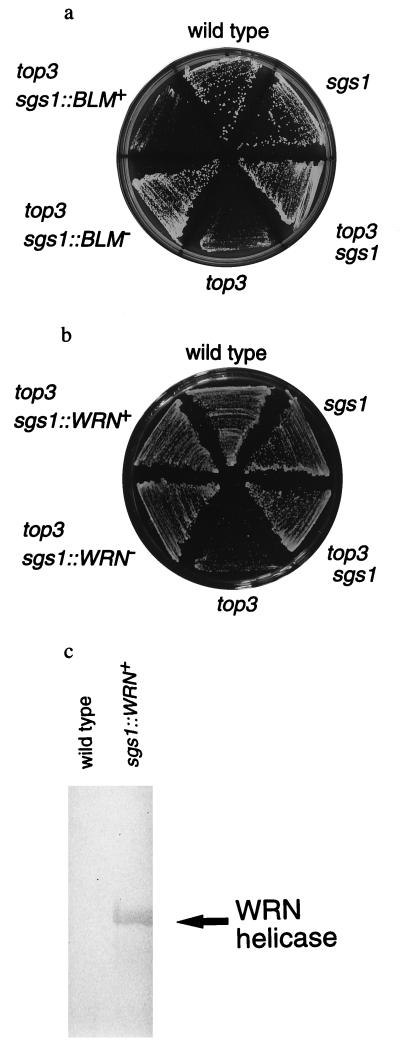
BLM Helicase Can Suppress the Growth in the top3 sgs1 Double Mutant but WRN Helicase Cannot.
To understand the other biological functions of BLM and WRN helicases, we examined whether BLM and WRN helicases can suppress the growth in the yeast top3 sgs1 double mutant. The top3 sgs1∷BLM+ double mutant showed the slow-growth phenotype, which is comparable to that in the top3 single mutant, whereas the top3 sgs1∷BLM− double mutant did not show the slow-growth phenotype (Fig. 5a). On the other hand, both top3 sgs1∷WRN+ and top3 sgs1∷WRN− double mutants did not show the slow-growth phenotype (Fig. 5b). Using Western blot analyses, we confirmed that WRN helicase is expressed in the sgs1∷WRN+ cells (Fig. 5c). The TOP3+ sgs1∷BLM+ and TOP3+ sgs1∷WRN+ strains grew almost as fast as the wild-type strain as described above. These results indicate that human BLM helicase can suppress the growth in the top3 sgs1 double mutant, but the WRN helicase cannot.
Figure 5.
Functions of BLM and WRN genes in the top3 background. (a) Comparison of the growth rates in the top3 sgs1∷BLM+ and top3 sgs1∷BLM− strains. We examined the growth rates of six strains: DH6.61D wild type, KY12 sgs1, KY15 top3 sgs1, KY16 top3, KY25 top3 sgs1∷BLM+, KY26 top3 sgs1∷BLM−. (b) Comparison of growth rates of the top3 sgs1∷WRN+ and top3 sgs1∷WRN− strains. We examined growth rates of six strains: DH6.61D wild type, KY12 sgs1, KY15 top3 sgs1, KY16 top3, KY27 top3 sgs1∷WRN+, and KY28 top3 sgs1∷WRN−. (c) Western blot analysis of DH6.61D wild type and KY23 sgs1∷WRN+. Western blot analysis of DH6.61D wild type and KY23 sgs1∷WRN+ was carried out as described in Materials and Methods.
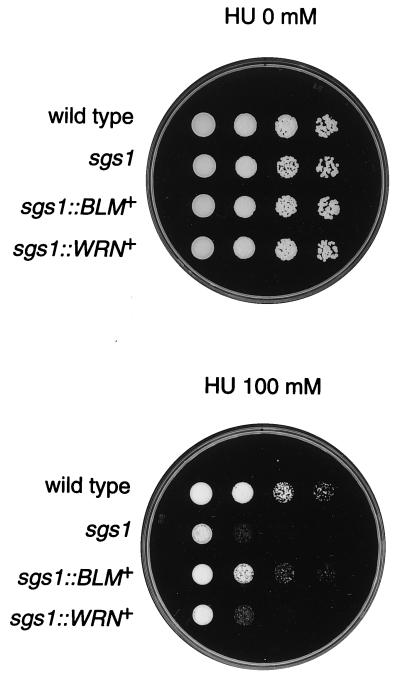
BLM Helicase Can Restore Hypersensitivity to Hydroxyurea in the sgs1 Mutant but WRN Helicase Cannot.
Because the sgs1 mutant is hypersensitive to hydroxyurea (HU) (T. Enomoto, personal communication), we examined whether BLM or WRN helicase can restore this particular phenotype in the sgs1 mutant. The sgs1∷BLM+ strain showed normal sensitivity to HU, as in the wild-type strain, whereas the sgs1∷WRN+ strain showed hypersensitivity to HU (Fig. 6). Both sgs1∷BLM− and sgs1∷WRN− also showed hypersensitivity to HU (data not shown). These results indicate that human BLM helicase can restore hypersensitivity to HU in the sgs1 mutant, but the WRN helicase cannot.
Figure 6.
Sensitivity of the sgs1∷BLM+ and sgs1∷WRN+ strains to hydroxyurea. Exponential cultures of DH6.61D wild type, KY12 sgs1, KY21 sgs1∷BLM+, and KY23 sgs1∷WRN+ were spotted onto synthetic complete plates with or without 100 mM hydroxyurea (HU). Numbers of cells per spot were 3 × 104, 3 × 103, 3 × 102, and 30 (from left to right).
DISCUSSION
In this paper, we showed that Sgs1 helicase suppresses illegitimate recombination in the YCpL2 plasmid system. Previously, Tsukamoto et al. (15, 16) indicated that Rad52 and Hdf1, which are involved in homologous recombination (28) and end-joining (16, 17), respectively, participate in deletions of the YCpL2 plasmid. This result can be explained by a model in which unstable dicentric plasmids formed by the Rad52-dependent homologous recombination between two replicated plasmids could be broken subsequently between two centromeres, resulting in deletions by the Hdf1-dependent end-joining. In fact, we indicated that the majority of illegitimate recombination enhanced by the sgs1 mutation depends on the Rad52 and Hdf1 functions and therefore is mediated by homologous recombination and end-joining processes. Furthermore, Sgs1 helicase was shown not to be involved in end-joining. These results are consistent with Tsukamoto’s model (18). Therefore, we infer that Sgs1 helicase has a function to suppress illegitimate recombination through regulating Rad52-dependent homologous recombination.
Gangloff et al. (12) and Watt et al. (9) have shown that homologous recombination is increased in the sgs1 mutant. The former indicated that the frequency of intramolecular recombination between rDNA repeats is enhanced by the sgs1 mutation. The latter showed that the frequencies of interchromosomal homologous recombination, intrachromosomal excision recombination, and ectopic recombination are increased by the sgs1 mutation. All these recombinations were carried out between extensive homologous sequences, but neither was carried out between short homologous sequences or nonhomologous sequences. In contrast, we showed that the frequency of illegitimate recombination is increased by the sgs1 mutation. It should be noted that the above experiments were carried out by using the YCpL2 plasmid system. But these results strongly suggest that the increase of illegitimate recombination through formation of a dicentric DNA molecule takes place not only on plasmids but also on chromosomes. Although we have shown that RecQ helicase works as a suppressor of illegitimate recombination in abnormal excision of chromosome in E. coli (10), a question whether Sgs1 can suppress illegitimate recombination in chromosome systems should be tested in yeast.
That the frequency of illegitimate recombination in the sgs1 rad52 mutant is higher than that in the rad52 single mutant indicates that the Rad52-independent illegitimate recombination also is increased by the sgs1 mutation, although the frequency of illegitimate recombination is much lower than that in the RAD52+ cell. A similar observation, that Rad52-independent homologous recombination was increased by the sgs1 mutation, was made by Watts et al. (9). It has not been clarified yet whether the minor illegitimate recombination pathway in the sgs1 rad52 genetic background takes place through Rad52-independent homologous recombination or whether this pathway takes place by illegitimate recombination not involving a homologous recombination process.
An increased instability of chromosome was observed in both BS and WS cells (1–3), and an increased rate of illegitimate or homologous recombination is thought to be a major cause of the instability in these cells. In this paper, both the BLM and WRN helicases were shown to have an activity that suppresses hyperrecombination in the sgs1 mutant. These results imply that BLM and WRN helicases have a role in regulating genomic stability through suppression of recombination in human cells.
In addition, BLM helicase can suppress the cell growth in the sgs1 top3 double mutant and can restore the hypersensitivity to hydroxyurea in the sgs1 mutant, although the WRN helicase does not carry these functions. These results may suggest that yeast Sgs1 helicase has functions similar to those of human BLM helicase. Lu et al. (29) examined the function of Sgs1 helicase in the top3 mutant background and found that the sgs1-hd mutation, the helicase-defective allele, can complement an sgs1 null mutant, but the sgs1-ct mutation, the C-terminal truncation, cannot. These findings suggest that the helicase activity is not the only function for this class of proteins. In our study, BLM helicase can compensate the function in the top3 mutant background, but the WRN helicase cannot. It therefore is thought that the N- or C-terminal domain of BLM helicase may be important for the function of growth suppression in the top3 mutation background and the corresponding domain of WRN helicase may differ from that of BLM helicase functionally. On the other hand, the helicase domain of BLM helicase may have the same function as that of WRN helicase and may be responsible for suppression of recombination.
Stewart et al. (30) reported that a mutation of the rqh1+ gene, recQ homologue in Schizosaccharomyces pombe, confers hypersensitivity to hydroxyurea. Our study indicated that BLM helicase can restore the increased sensitivity of the sgs1 mutant to hydroxyurea. These results imply that Sgs1, Rqh1, and BLM helicases have roles for resumption of cell cycle progression when S phase has been interrupted.
Recently the sgs1 mutant was found to show a phenotype of premature aging in yeast mother cells with a reduced lifespan (13), which suggests that Sgs1 helicase also has additional functions related to cellular senescence, corresponding to the functions of the WRN helicase in humans. Sinclair and Guarante (14) showed that extrachromosomal rDNA circles (ERCs) are accumulated in old cells and the mutation in the SGS1 gene enhances the accumulation of ERCs, suggesting that it leads to premature aging and a shorter lifespan. Our findings indicate that both the BLM and WRN helicases can suppress the hyperrecombination in the sgs1 mutant. In view of the cells from the WS patients, but not the BS patients, which exhibit a shorter lifespan, the suppression of hyperrecombination by WRN helicase per se may not relate directly to a shorter lifespan. It is worth examining whether the WRN helicase, but not BLM, prevents early aging in yeast sgs1 mutant reported by Sinclair et al. (13). This possibility can be studied in our yeast system.
Acknowledgments
We thank Drs. A. Sugino and A. Miyajima for plasmids, Y. Tsukamoto for critical reading of the manuscript, and T. Enomoto for personal communication before publication. This work was supported in part by grants to J.K. and H.I. from the Ministry of Education, Science, Sports, and Culture of the Japan. This work was also supported by the Organization for Drug ADR Relief, R & D Promotion, and Product Review of the Japanese Government.
ABBREVIATIONS
- BS
Bloom’s syndrome
- WS
Werner’s syndrome
- HU
hydroxyurea
References
- 1.Langlois R G, Bigbee W L, Jensen R H, German J. Proc Natl Acad Sci USA. 1989;86:670–674. doi: 10.1073/pnas.86.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scappaticci S, Cerimele D, Fraccaro M. Hum Genet. 1982;62:16–24. doi: 10.1007/BF00295599. [DOI] [PubMed] [Google Scholar]
- 3.Fukuchi K, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monnat R J, Jr, Hackmann A F, Chiaverotti T A. Genomics. 1992;13:777–787. doi: 10.1016/0888-7543(92)90153-j. [DOI] [PubMed] [Google Scholar]
- 5.Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 6.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 8.Umezu K, Nakayama K, Nakayama H. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watt P M, Hickson I D, Borts R H, Louis E J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt P M, Louis E J, Borts R H, Hickson I D. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 12.Gangloff S, McDonald J P, Bendxien C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair D A, Mills K, Guarente L. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair D A, Guarente L. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 15.Tsukamoto Y, Kato J, Ikeda H. Genetics. 1996;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto Y, Kato J, Ikeda H. Nucleic Acids Res. 1996;24:2067–2072. doi: 10.1093/nar/24.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milne G T, Jin S, Shannon K B, Weaver D T. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukamoto Y, Kato J, Ikeda H. Mol Gen Genet. 1997;255:543–547. doi: 10.1007/s004380050527. [DOI] [PubMed] [Google Scholar]
- 19.Schiestl R H, Zhu J, Petes T D. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukamoto Y, Kato J, Ikeda H. Nature (London) 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 22.Wilson T E, Grawunder U, Lieber M R. Nature (London) 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 23.Teo S-H, Jackson S P. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schär P, Herrmann G, Daly G, Lindahl T A. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 26.Broach J R, Strathern J N, Hicks J B. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 27.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugawara N, Ivanov E L, Fishman-Lobell J, Ray B L, Wu X, Haber J E. Nature (London) 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglantz R. Nature (London) 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 30.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]