Abstract
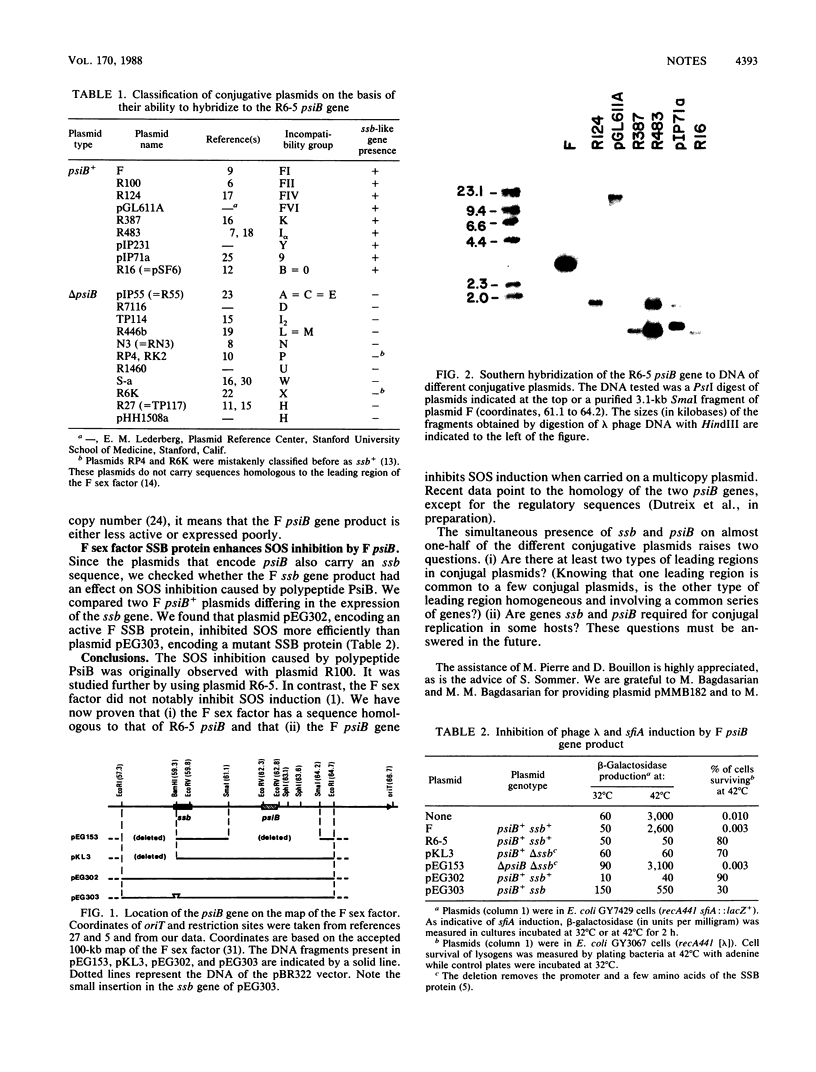
In 9 of 20 conjugative plasmids of different incompatibility groups, including F and R100 (or R6-5), coexist two sequences which are homologous, respectively, to the gene psiB, which encodes an inhibitor of SOS induction, and to the gene ssb, which encodes a single-stranded-DNA-binding protein.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Bailone A., Bagdasarian M. M., Manning P. A., Lurz R., Timmis K. N., Devoret R. An inhibitor of SOS induction, specified by a plasmid locus in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5723–5726. doi: 10.1073/pnas.83.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., D'Ari R., Filipowicz W., George J. Suppression of induction of SOS functions in an Escherichia coli tif-1 mutant by plasmid R100.1. J Bacteriol. 1980 Feb;141(2):464–469. doi: 10.1128/jb.141.2.464-469.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailone A., Blanco M., Devoret R. E. coli K12 inf: a mutant deficient in prophage lambda induction and cell filamentation. Mol Gen Genet. 1975;136(4):291–307. doi: 10.1007/BF00341714. [DOI] [PubMed] [Google Scholar]
- Castellazzi M., George J., Buttin G. Prophage induction and cell division in E. coli. I. Further characterization of the thermosensitive mutation tif-1 whose expression mimics the effect of UV irradiation. Mol Gen Genet. 1972;119(2):139–152. doi: 10.1007/BF00269133. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Merrill B. M., Williams K. R. F sex factor encodes a single-stranded DNA binding protein (SSB) with extensive sequence homology to Escherichia coli SSB. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5480–5484. doi: 10.1073/pnas.80.18.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Barth P. T. Compatibility properties of R483, a member of the I plasmid complex. J Bacteriol. 1976 Mar;125(3):796–799. doi: 10.1128/jb.125.3.796-799.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Compatibility groups among fi - R factors. Nature. 1971 Nov 26;234(5326):222–223. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Host ranges of R factors. J Gen Microbiol. 1972 May;70(3):453–460. doi: 10.1099/00221287-70-3-453. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Olarte J. R factors in strains of Salmonella typhi and Shigella dysenteriae 1 isolated during epidemics in Mexico: classification by compatibility. Antimicrob Agents Chemother. 1974 Mar;5(3):310–317. doi: 10.1128/aac.5.3.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J., Galindo E., Olarte J., Falkow S. Beta-lactamase of R factors. J Bacteriol. 1968 Oct;96(4):1441–1442. doi: 10.1128/jb.96.4.1441-1442.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Conjugative plasmids of enteric bacteria from many different incompatibility groups have similar genes for single-stranded DNA-binding proteins. J Bacteriol. 1985 Apr;162(1):235–241. doi: 10.1128/jb.162.1.235-241.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Unrelated conjugative plasmids have sequences which are homologous to the leading region of the F factor. J Bacteriol. 1986 May;166(2):670–672. doi: 10.1128/jb.166.2.670-672.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Grindley J. N., Anderson E. S. R factor compatibility groups. Mol Gen Genet. 1972;119(4):287–297. doi: 10.1007/BF00272087. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Coetzee J. N., Dennison S. R factors from Proteus morganii. J Gen Microbiol. 1973 Aug;77(2):249–259. doi: 10.1099/00221287-77-2-249. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N. Plasmids determining I pili constitute a compatibility complex. J Gen Microbiol. 1973 Jul;77(1):19–25. doi: 10.1099/00221287-77-1-19. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N. R124, an fi R factor of a new compatibility class. J Gen Microbiol. 1972 Jul;71(2):403–405. doi: 10.1099/00221287-71-2-403. [DOI] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. Effect of suppressors of SOS-mediated filamentation on sfiA operon expression in Escherichia coli. J Bacteriol. 1983 Jan;153(1):169–175. doi: 10.1128/jb.153.1.169-175.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin A. L., Capage M. A., Golub E. I., Low K. B. F sex factor of Escherichia coli K-12 codes for a single-stranded DNA binding protein. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4422–4426. doi: 10.1073/pnas.80.14.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontomichalou P., Mitani M., Clowes R. C. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J Bacteriol. 1970 Oct;104(1):34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M., Hedges R. W. Analytical isoelectric focusing of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1976 Feb;125(2):713–718. doi: 10.1128/jb.125.2.713-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- Ray A., Skurray R. Cloning and polypeptide analysis of the leading region in F plasmid DNA transfer. Plasmid. 1983 May;9(3):262–272. doi: 10.1016/0147-619x(83)90004-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sommer S., Bailone A., Devoret R. SOS induction by thermosensitive replication mutants of miniF plasmid. Mol Gen Genet. 1985;198(3):456–464. doi: 10.1007/BF00332939. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Furuse C., Sakaizumi S. Transduction of various R factors by phage P1 in Escherichia coli and by phage P22 in Salmonella typhimurium. J Bacteriol. 1968 Nov;96(5):1791–1795. doi: 10.1128/jb.96.5.1791-1795.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. From Gainesville to Toulouse: the evolution of a model. Biochimie. 1982 Aug-Sep;64(8-9):549–555. doi: 10.1016/s0300-9084(82)80086-2. [DOI] [PubMed] [Google Scholar]