Abstract
The human β-globin locus control region (LCR), which consists of four erythroid-specific DNase I hypersensitive sites (HS1–HS4), functions over a long distance to control the transcription, chromatin structure, and replication of the β-globin genes. We have used stable transfection assays to show that activation of the mitogen-activated protein (MAP) kinase pathway by low concentrations of the phorbol ester phorbol 12-tetradecanoate 13-acetate (TPA) induces enhancer activity of the LCR subregion HS2, but not HS3. Although HS2 enhancer activity is diminished with increasing distance from the promoter, the relative level of induction by TPA is independent of HS2–promoter distance. Mutation of cis-elements within HS2 reveals that the tandem-binding sites for the hematopoietic-specific transcription factor NF-E2 are required for induction by TPA, and induction is conferred by expressing NF-E2 in an NF-E2-null cell line. These results show that MAP kinases target factors functioning through the NF-E2 sites to enhance long-range transactivation by the LCR.
In higher eukaryotes, transcriptional regulatory elements, such as enhancers and locus control regions (LCRs), often are located thousands of base pairs away from the genes they influence. A common feature to enhancers and LCRs is the presence of clustered binding sites for transcription factors. Protein–protein interactions between promoter-bound transcription factors and components of the basal transcription machinery are known to be important for activation. However, the mechanism by which transcription factors bound to distal elements confer long-range activation is largely unsolved. Several mechanisms have been proposed to explain long-range activation (reviewed in ref. 1). A looping model suggests that factors bound to the distal elements physically interact with promoter-bound factors, with concomitant looping of the intervening DNA. As a result, the basal transcription machinery is recruited to the promoter. A scanning or tracking mechanism assumes that transcription factors binding to distal regulatory sites move or track processively toward the promoter (2). In this model, either the distal factors could recruit RNA polymerase once they have reached the promoter, or RNA polymerase could be recruited to the distal site and then migrate toward the promoter as a complex with the transacting factor. A third model predicts that factors bound to the distal sites are necessary for modulating the chromatin structure of the promoter to increase accessibility for the transcriptional machinery (3–6).
We have been using the human β-globin LCR as a paradigm to study mechanisms of long-range activation. The LCR functions over a long distance (>50 kb) to control the transcription, chromatin structure, and replication of the β-globin genes (7–9). In addition, the LCR confers high level, position-independent, and copy number-dependent expression on linked globin transgenes in mice (9). The importance of the LCR is also evident from naturally occurring deletions that remove LCR sequences, resulting in severe β-thalassemia, even though the globin genes are intact (7).
The LCR consists of four erythroid-specific DNase I hypersensitive sites (HS1–HS4) (10). HS2 and HS3 each have strong enhancer activities in stable transfection assays and transgenic mice (4, 11–15). HS4 also has moderate enhancer activity, but HS1 appears to lack enhancer activity (16). We have shown previously that HS2 and HS3 have a limited functional distance for their enhancer properties, whereas an LCR containing all four HSs retains strong enhancer activity at a distance of 7.3 kb (5). This suggests that synergistic interactions between HSs are required for long-range activation. We have proposed that these interactions create a stable nucleoprotein complex that recruits chromatin-modifying enzymes, and that these enzymes mediate the effects of the LCR on chromatin structure (5, 6). Although signaling mechanisms necessary for erythropoiesis have been well studied (17), the impact of these mechanisms on LCR function are unknown. Therefore, we have investigated the possible involvement of protein kinase C (PKC) and mitogen-activated protein (MAP) kinases in the long-range function of the LCR. In this paper, we show that PKC activation of the MAP kinase signaling pathway by low concentrations of the phorbol ester phorbol 12-tetradecanoate 13-acetate (TPA) induces HS2 enhancer activity, independent of HS2-promoter distance. We also demonstrate the importance of tandem-binding sites within HS2 for the hematopoietic transcription factor NF-E2 and implicate NF-E2 as a factor targeted by this signaling cascade.
MATERIALS AND METHODS
Cell Culture.
The human erythroleukemia cell line K562 was propagated as described previously (5). CB3 cells, obtained from Y. Ben-David (Univ. of Toronto), were propagated in DMEM containing 10% FBS/2 mM glutamine/25 μg/ml gentamycin/1% antibiotic-antimycotic solution (GIBCO/BRL). All cell lines were grown in a humidified incubator at 37°C, in the presence of 5% CO2. Stably transfected pools or clones of K562 cells were selected and maintained in the presence of 0.2 mg/ml and 0.1 mg/ml hygromycin B, respectively. Stably transfected pools of CB3 cells were selected and maintained in the presence of 1 mg/ml active G418.
Plasmid Construction.
Construction of the vectors pγluc, pGL3RIγluc, pHS2γluc, pHS3γluc, pminiLCRγluc, and derivatives of these vectors containing various lengths of phage λ DNA subcloned upstream of the γ-globin promoter was described previously (5). Each of these vectors contains the luciferase reporter gene of pGL3basic (Promega) driven by the human γ-globin promoter (−260 to +35). In this report, HS2 refers to the KpnI (7768)–BglII (9218) human HS2 fragment, HS3 refers to the HindIII (3266)–HindIII (5172) human HS3 fragment, and miniLCR refers to an EcoRI–SalI fragment (4) containing the conserved core regions of human HS1–HS4 and natural flanking sequences (coordinates: HS1, 10946–15180; HS2, 7764–9218; HS3, 4277–5122; HS4, 951–2199). A 1,097-bp KpnI–XbaI fragment of pHS2γluc, containing the 5′ region of HS2, and a 750-bp XbaI–HindIII fragment of pA119 (provided by J. Cunningham, St. Jude Children’s Hospital, Memphis, TN), containing the complementing 3′ region of HS2 and the β-globin promoter, were subcloned into pGL3basic to yield pHS2βluc. Integrity of plasmids was confirmed by restriction enzyme digestion analysis.
Stable Transfections.
K562 cells were cotransfected by electroporation as described (5), using linearized test (5 μg) and selection (0.5 μg) plasmids. Test and selection plasmids were linearized at unique NotI and SalI sites, respectively. CB3 cells were cotransfected similarly, but 10 μg of linearized test plasmid and 2.5 μg of a linearized selection plasmid, conferring resistance to G418, were used. Test and selection plasmids for CB3 transfections were linearized at unique NotI sites. The NF-E2 expression plasmid, pEF1αneo45-G1 (18), was treated as a selection plasmid (2.5 μg), and was linearized at a unique BsaI site.
Luciferase Assays.
Cells (1.4 ml of a near-confluent culture) were isolated by centrifugation at 240 × g for 5 min at 4°C and washed by resuspension in 0.5 ml of ice-cold PBS and recentrifugation. The cells were lysed in 40 μl of reporter lysis buffer (Promega) for 15 min at room temperature, and the supernatant was isolated after centrifugation for 5 min at 18,700 × g. Luciferase activity generated by the supernatant in 30 s was determined with a Berthold Lumat LB9501 luminometer. Luciferase values were normalized by protein concentration, which was estimated by the Bradford assay using γ-globulin as a standard.
RESULTS AND DISCUSSION
Transactivation by HS2 Is Enhanced by TPA, Independent of HS2-Promoter Distance.
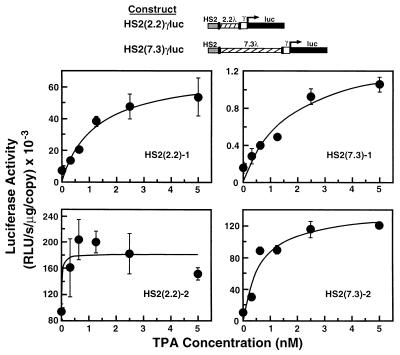
The phorbol ester TPA has been shown to stimulate the MAP kinase pathway via activation of PKC (19, 20). To investigate the possible involvement of PKC and MAP kinases in long-range activation by the LCR, we examined the effect of TPA on K562 clonal cell lines, stably transfected with constructs containing HS2 separated from a γ-globin promoter/luciferase reporter by either 2.2 or 7.3 kb of phage λ DNA. We observed that TPA treatment induced luciferase reporter activity with maximal induction occurring with 5 nM TPA and within 20 h (Fig. 1). Interestingly, the magnitude of induction was relatively constant (2- to 6-fold) regardless of whether HS2 was located near or far from the promoter. This suggests that a TPA-induced signaling mechanism enhances long-range activation by HS2.
Figure 1.
TPA induces luciferase reporter activity independent of the distance between HS2 and the promoter. K562 clonal cell lines, stably transfected with either of the constructs shown, were treated with TPA for 20 h, and then luciferase activity was determined from cell lysates as a measure of γ-globin promoter activity (mean ± SEM). At least triplicate measurements were made for each TPA concentration. The copy number of integrated templates was 8, 2, 22, and 1 for clones HS2(2.2)-1, HS2(2.2)-2, HS2(7.3)-1, and HS2(7.3)-2, respectively.
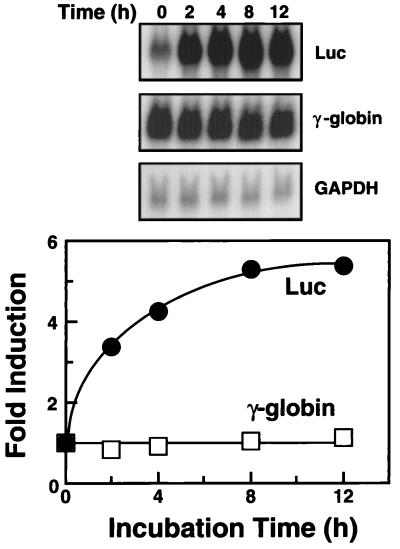
To determine whether TPA induction occurs at the level of mRNA, we measured steady-state luciferase mRNA in the stably transfected clonal cell line HS2(2.2)-1 (see Fig. 1), treated with 5 nM TPA for up to 12 h (Fig. 2). The level of luciferase mRNA increased rapidly and then plateaued at 8–12 h. Because the degree of induction of luciferase mRNA (5.4-fold at 12 h) and activity was similar, luciferase mRNA accumulation appears to account for the TPA effect.
Figure 2.
TPA induction of luciferase mRNA. Total RNA isolated from the stably transfected clonal cell line HS2(2.2)-1, treated with 5 nM TPA for the indicated time, was examined by Northern blot analysis. Shown are the results of successive hybridization of the same filter to the luciferase, γ-globin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene probes. Signal intensities were quantified by using a PhosphorImager, and luciferase and γ-globin mRNA levels, normalized to those of GAPDH, were plotted.
K562 cells can be induced with TPA to differentiate into megakaryocytes (21). However, it should be noted that our conditions for TPA induction do not induce significant megakaryocytic differentiation, because endogenous γ-globin mRNA levels, which are down-regulated at an early stage of differentiation (22), were unaffected (Fig. 2). This does not rule out the possibility that some initial steps in the differentiation pathway occur during the TPA treatment. All cells underwent megakaryocytic differentiation after treatment with 50 nM TPA for 48–72 h, indicating that the cells were competent in this well characterized developmental pathway.
HS2 Mediates TPA Induction.
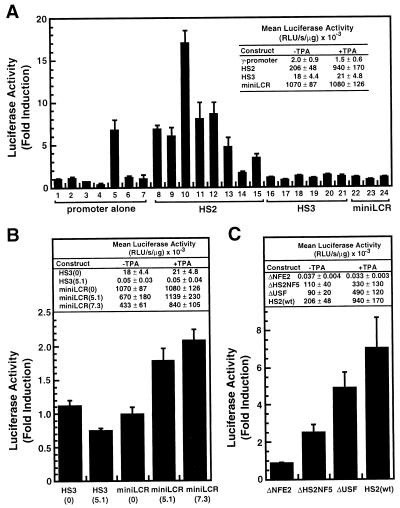
To examine whether induction is mediated by HS2, we analyzed pools of stably transfected cells containing γ-globin promoter/luciferase reporter constructs, in the absence or presence of HS2, HS3, and the miniLCR. These constructs lack phage λ DNA inserts. Although all pools had luciferase activity, only the HS2γluc construct was induced consistently by TPA (Fig. 3A). This rules out the possibility that TPA causes a general increase in luciferase mRNA stability and argues against a TPA-induced change in the activity of a general transcription factor, RNA polymerase, or a permissive modification of chromatin, which would likely affect all of the constructs.
Figure 3.
NF-E2 sites within HS2 are required for TPA induction. (A) Stably transfected pools of K562 cells containing either the γluc, HS2γluc, HS3γluc, or miniLCRγluc constructs were treated with either 5 nM TPA or vehicle for 20 h and then analyzed for luciferase activity as a measure of γ-globin promoter activity. The fold induction for each pool (mean ± SEM, triplicate measurements) is shown plotted, and the mean activity ± SEM for each construct is shown in the inset. (B) Stably transfected pools containing either HS3γluc (n = 6), HS3(5.1)γluc (n = 3), miniLCRγluc (n = 3), miniLCR(5.1)γluc (n = 4), or miniLCR(7.3)γluc (n = 4) constructs were analyzed for the effect of TPA as above. The mean fold induction ± SEM is shown plotted, and the mean activity ± SEM is shown in the Inset. (C) Stably transfected pools containing either ΔNF-E2 (n = 10), ΔHS2NF5 (n = 4), ΔUSF (n = 4), or HS2γluc (n = 8) constructs were analyzed as above. The mean fold induction ± SEM is shown plotted, and the mean activity ± SEM is shown in the Inset. At least triplicate measurements were made for each sample.
The consistent induction of HS2γluc indicates that this construct contains all the cis-acting elements necessary for induction. Because HS2γluc shares identical vector, promoter, and reporter sequences with all of the constructs tested, and the same selection plasmid was used in each transfection, the HS2 region must be the critical cis-acting element. An ability to augment the activity of any enhancer is unlikely, because both HS3 and the miniLCR clearly enhance reporter activity (see Fig. 3A, Inset).
The absence of TPA induction with the miniLCRγluc construct seems paradoxical, because the miniLCR contains HS2. However, the miniLCR, when located near the promoter, may already activate transcription at a maximal capacity and cannot be induced further. This maximal activity may reflect the ability of the miniLCR to efficiently form a decondensed chromatin domain, similar to the endogenous LCR (5, 7, 9). We postulate that the effects of TPA influence the formation, but not the maintenance of the active domain. The lack of TPA induction of endogenous γ-globin mRNA levels (Fig. 2) is consistent with this idea.
The enhancer activity of the miniLCR can be reduced severalfold by increasing its distance from the promoter (5). Under these conditions, the less-than-maximally active miniLCR may be sensitive to the effects of TPA, mediated by HS2. To test this hypothesis, we examined the effect of TPA on pools of K562 cells, stably transfected with constructs containing the miniLCR separated from the promoter by either 5.1 or 7.3 kb of phage λ DNA. The miniLCR(5.1)γluc and miniLCR(7.3)γluc constructs both were induced by TPA, 1.8- and 1.9-fold, respectively (Fig. 3B). Pools transfected with a construct containing HS3 placed 5.1 kb upstream from the promoter, remained insensitive to TPA, indicating that phage λ DNA sequences are not responsible for induction. The induction of distance-constrained miniLCR constructs, but not constructs in which the miniLCR is located near the promoter, suggests that TPA treatment increases the effective distance over which the miniLCR can function in a chromatin environment.
One of seven pools stably transfected with the γluc construct was induced by TPA to a level (6.7-fold) similar to that observed with HS2γluc (lane 5, Fig. 3A). However, no other pool transfected with γluc, HS3γluc, or miniLCRγluc was induced, indicating that induction is not mediated solely by the γ-globin promoter. Therefore, it seems likely that the one inducible γluc pool contains the γluc construct integrated near a TPA-responsive element.
NF-E2 Binding Sites Within HS2 Are Required for TPA Induction.
It is possible that no single element of HS2 is responsible for TPA induction, but rather a combination of sites, as multiple factors contribute to the enhancer activity of HS2 (23). To delineate cis-acting elements necessary for induction, we evaluated the effect of TPA in stable transfection assays, in which the HS2 element of pHS2γluc was substituted with mutated HS2 elements, each containing a single deletion of factor-binding sites known to be important for enhancer activity (Fig. 3C). Mutations that preclude binding of either HS2NF5 (ΔHS2NF5, positions 8701–8706; CCCAGATGTT changed to CCGTCGACTT), or USF (ΔUSF, positions 8790–8795; ACCACCTGAC changed to ACGTCGACAC), inhibit enhancer activity by 46.6 and 56.3%, respectively. Similar levels of inhibition have been reported for these mutations (23, 24). Both of these mutant constructs were induced by TPA, indicating that full HS2 enhancer activity is not required for induction. However, multiple HS2-binding factors may influence the efficiency of TPA induction, because the average level of induction observed with the ΔHS2NF5 and ΔUSF constructs is less than that of the wild-type HS2 construct.
The tandem NF-E2 sites within HS2 are known to be critical for enhancer activity (13, 23, 24). Mutation of the NF-E2 binding sites (ΔNF-E2, positions 8661–8677; GCTGAGTCATGATGAGTCATG changed to GCGTCGACTG) resulted in low, but reproducible reporter activity, consistent with the known importance of this site for high activity in transfection assays and in transgenic mice. However, unlike the pools transfected with ΔHS2NF5 and ΔUSF constructs, no TPA induction was observed in any of 10 pools stably integrated with the ΔNF-E2 construct, indicating that NF-E2 sites are required for induction (Fig. 3C).
The absence of induction in pools stably transfected with the ΔNF-E2 construct suggests that proteins binding to these sites may mediate induction by TPA. In addition to NF-E2, other proteins have been shown to bind these sites, including AP-1, LCR-F1/Nrf-1, Nrf-2, Bach1, and Bach2 (25–29). However, the role of these proteins in LCR function is unknown.
NF-E2 has been shown to be critical for globin gene expression in mouse erythroleukemia CB3 cells. CB3 cells do not express the p45 subunit of NF-E2 because of proviral integration in one allele and loss of the second allele (30). The lack of p45 results in extremely reduced levels of globin gene expression, which can be restored by introduction of NF-E2 (18, 31–33), indicating that other factors that bind the target sites are insufficient for activation in these cells. We used this model system to determine whether NF-E2 is necessary for induction by TPA. We examined the effect of TPA on pools of CB3 cells, stably transfected with the HS2βluc plasmid, in the presence and absence of a stably integrated NF-E2 expression vector, pEF1αneo45-G1 (18). Because CB3 cells are commonly treated with dimethyl sulfoxide (DMSO) to induce erythroid differentiation, we examined cells with and without DMSO treatment. As shown in Table 1, CB3 pools expressing NF-E2 had a 10-fold greater luciferase activity than pools without NF-E2, regardless of whether the cells were treated with DMSO. This indicates that sufficient amounts of NF-E2 are produced in these transfected pools to restore HS2 enhancer activity of the integrated construct. In addition, only the NF-E2-expressing pools were inducible with TPA (−DMSO, P = 0.09, n = 3; +DMSO, P = 0.03, n = 3); pools without NF-E2 were uninduced. Thus, NF-E2 is required for induction by TPA, and other factors that can bind to the NF-E2 sites in these cells do not share this property. The lower level of induction in CB3 cells, compared with those observed with HS2γluc and K562 cells, may reflect the overexpression of NF-E2 in this system or a requirement for additional components for a maximal response.
Table 1.
NF-E2 is required for TPA induction
| Cells | DMSO treatment | Average luc activity, RLU/s per μg
|
Average fold induction | |
|---|---|---|---|---|
| −TPA | +TPA | |||
| CB3 | No | 440 ± 240 | 450 ± 240 | 1.04 ± 0.02 |
| CB3 + NF-E2 | No | 4,580 ± 1,580 | 6,780 ± 2,840 | 1.41 ± 0.12 |
| CB3 | Yes | 700 ± 290 | 690 ± 300 | 0.98 ± 0.04 |
| CB3 + NF-E2 | Yes | 7,400 ± 1,900 | 10,000 ± 3,400 | 1.30 ± 0.13 |
Stably transfected pools of CB3 cells containing either HS2βluc (n = 3) or HS2βluc and the NF-E2 expression vector pEF1αneo45-G1 (n = 3) were treated with either 5 nM TPA or vehicle for 20 h, both in the presence and absence of 1.8% dimethyl sulfoxide (DMSO), and then analyzed for luciferase activity as a measure of β-globin promoter activity (mean ± SEM). At least triplicate measurements were made for each sample.
Involvement of the MAP Kinase Pathway in TPA Induction.
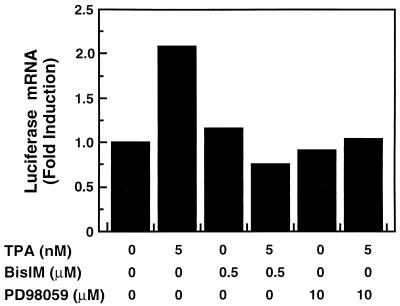
TPA has been shown to stimulate the MAP kinase pathway via direct activation of PKC (19, 20). To determine whether TPA acts through this pathway in our system, we determined whether two inhibitors of this pathway influence TPA induction of luciferase mRNA. K562 cells, stably transfected with the HS2γluc construct, were treated as indicated in Fig. 4, and the steady-state levels of luciferase mRNA were determined by Northern analysis. After a 2-h exposure to 5 nM TPA, luciferase mRNA levels were increased 2.1-fold. However, addition of either the PKC inhibitor bisindolylmaleimide or the MEK1 inhibitor PD98059 30 min before addition of TPA inhibited induction (lanes 4 and 6). Neither inhibitor significantly affected luciferase mRNA levels in the absence of TPA (lanes 3 and 5). Therefore, we conclude that induction of HS2 enhancer activity by TPA is the result of PKC activation of the MAP kinase signaling pathway. During the review of this manuscript, it was reported that MAP kinases facilitate transactivation by NF-E2 from synthetic reporter constructs, consistent with our results with HS2 and the miniLCR (34).
Figure 4.
TPA induction is blocked by PKC and MEK1 inhibition. Total RNA isolated from the stably transfected clonal cell line HS2(2.2)-1, treated as indicated, was analyzed by Northern blotting. Cells were exposed to inhibitors or vehicle for 2.5 h. For those treatments in which TPA was included, TPA was present during the final 2 h of the treatment. Luciferase mRNA levels were quantified and normalized to those encoding glyceraldehyde-3-phosphate dehydrogenase.
The involvement of MAP kinase signaling in hematopoiesis is complex. Activation of this pathway is required for both megakaryocytic differentiation (20, 35) and erythropoietin-induced erythroid differentiation and proliferation (36, 37). The basis for these opposing effects of MAP kinases is unknown. However, it seems likely that critical differences exist in the kinetics of down-regulation and specificity of the PKC isoforms required for MAP kinase activation (38).
Implications of MAP Kinase Signaling for Long-Range Activation by the LCR.
Previously, we proposed that long-range activation by the β-globin LCR requires recruitment of chromatin-modifying enzymes, and that these enzymes mediate the effects of the LCR on chromatin structure (5, 6, 39). Here, we have shown that activation of the MAP kinase pathway enhances long-range transactivation by HS2, that the tandem NF-E2-binding sites of HS2 are required for induction, and that induction is conferred by NF-E2 in an NF-E2-null cell line. Because the p45 subunit of NF-E2 is a substrate for diphosphorylated ERK2 in vitro (E. A. Mosser, E. C. Forsberg, N. G. Ahn, A. Vassilev, and E.H.B., unpublished results), we propose that phosphorylation of NF-E2, and perhaps other proteins that bind to the NF-E2 sites, facilitates recruitment of coactivators necessary for long-range transactivation.
Several proteins have been suggested to have a role in activation by NF-E2, based on their ability to interact with the activation domain of p45. Cheng et al. (40) have shown that p45 physically interacts with a glutathione S-transferase fusion protein containing a fragment of the transcriptional coactivator CBP/p300. In addition, we have shown that conserved residues within the activation domain of p45 mediate CBP/p300 binding, and CBP/p300 potentiates transactivation by HS2 in transient transfection assays only when NF-E2 sites are present (E. A. Mosser, E. C. Forsberg, N. G. Ahn, A. Vassilev, and E.H.B., unpublished results). CBP/p300 has histone acetyltransferase (HAT) activity (41), which may enhance transactivation through acetylation of histone amino-terminal tails. CBP/p300 also interacts with another HAT, P/CAF (42), which is present in a large macromolecular complex in globin-expressing, human K562 erythroleukemia cells (43).
Gavva et al. (44) identified several WW domain-containing proteins that interact with p45, including hRPF1 and its yeast homolog, RSP5. hRPF1 potentiates transactivation by human glucocorticoid and progesterone receptors in yeast and mammalian cells (45). The yeast and murine homologs of hRPF1, NEDD4, and RSP5, respectively, each possess ubiquitin ligase activity (46, 47). We have identified a human ubiquitin ligase homolog, WWP1, which binds sequences within the transactivation domain of p45, distinct from those necessary for CBP/p300 binding (E. A. Mosser, J. D. Kasanov, E. C. Forsberg, B. K. Kay, P. Ney, and E.H.B., unpublished results). In addition, Amrolia et al. (31) have shown that TAFII130, a component of the TFIID multiprotein complex, interacts with the p45 activation domain by yeast two-hybrid and glutathione S-transferase fusion assays.
Although NF-E2 is necessary for globin expression in CB3 cells and in transfection assays, p45 null mice exhibit only a mild deficit in globin expression (49), suggesting that other proteins binding to the NF-E2 sites or another region of the LCR may have a functionally redundant role in mice. Recruitment of a common set of coactivators by these proteins may explain the redundancy. In support of this idea, both AP-1 (50) and the hematopoietic transcription factor GATA-1 (51) bind CBP/p300. Both of these proteins also are targeted by phosphorylation (39, 48). Multisite phosphorylation regulates both DNA binding and transactivation by AP-1, whereas the consequences of GATA-1 phosphorylation are unknown. Our results implicate MAP kinase signaling in facilitating long-range transactivation by the LCR, suggesting that LCR activity may be modulated by such a signaling pathway during hematopoiesis.
Acknowledgments
We acknowledge support from the Milwaukee Foundation, the Leukemia Society of America, the Hemophilia Association of New York (Grant 133BK04), and the National Institutes of Health (Grant DK50107). E.H.B. is a Leukemia Society of America Scholar and a Shaw Scientist.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PKC, protein kinase C; MAP, mitogen-activated protein; LCR, locus control region; HS, hypersensitive site; TPA, phorbol 12-tetradecanoate 13-acetate.
References
- 1.Schlief R. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 2.Ouhammouch M, Sayre M H, Kadonaga J T, Geiduschek E P. Proc Natl Acad Sci USA. 1997;94:6718–6723. doi: 10.1073/pnas.94.13.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsenfeld G. Nature (London) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 4.Forrester W C, Novak U, Gelinas R, Groudine M. Proc Natl Acad Sci USA. 1989;86:5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnick E H, Tze L. Proc Natl Acad Sci USA. 1997;94:4566–4571. doi: 10.1073/pnas.94.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnick E H. Chemtracts-Biochem Mol Biol. 1997;10:139–147. [Google Scholar]
- 7.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 8.Forrester W C, Thompson C, Elder J T, Groudine M. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 10.Tuan D, London I M. Proc Natl Acad Sci USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon A M, Ley T J. Blood. 1991;77:2272–2284. [PubMed] [Google Scholar]
- 12.Hardison R, Xu J, Jackson J, Mansberger J, Selifonova O, Grotch B, Biesecker J, Petrykowska H, Miller W. Nucleic Acids Res. 1993;21:1265–1272. doi: 10.1093/nar/21.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talbot D, Philipsen S, Fraser P, Grosveld F. EMBO J. 1990;9:2169–2177. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philipsen S, Talbot D, Fraser P, Grosveld F. EMBO J. 1990;9:2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtin P T, Liu D P, Liu W, Chang J C, Kan Y W. Proc Natl Acad Sci USA. 1989;86:7082–7086. doi: 10.1073/pnas.86.18.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruzina S, Hanscombe O, Whyatt D, Grosveld F, Philipsen S. Nucleic Acids Res. 1991;19:1413–1419. doi: 10.1093/nar/19.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smithgall T E. Pharmacol Rev. 1998;50:1–19. [PubMed] [Google Scholar]
- 18.Blank V, Kim M J, Andrews N C. Blood. 1997;89:3925–3935. [PubMed] [Google Scholar]
- 19.Nishizuka Y. Science. 1986;233:305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- 20.Whalen A M, Galasinski S C, Shapiro P S, Nahreini T S, Ahn N G. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabilio A, Pelicci P G, Vinci G, Mannoni P, Civin C I, Vainchenker W, Testa U, Lipinski M, Rochant H, Breton-Gorius J. Cancer Res. 1983;43:4569–4574. [PubMed] [Google Scholar]
- 22.Lumelsky N L, Forget B G. Mol Cell Biol. 1991;11:3528–3536. doi: 10.1128/mcb.11.7.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caterina J J, Ciavatta D J, Donze D, Behringer R R, Townes T M. Nucleic Acids Res. 1994;22:1006–1011. doi: 10.1093/nar/22.6.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam L T, Bresnick E H. J Biol Chem. 1996;271:32421–32429. doi: 10.1074/jbc.271.50.32421. [DOI] [PubMed] [Google Scholar]
- 25.Caterina J J, Donze D, Sun C, Ciavatta D J, Townes T M. Nucleic Acids Res. 1994;1994:2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan J Y, Han X L, Kan Y W. Proc Natl Acad Sci USA. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moi P, Chan K, Asunis I, Cao A, Kan Y W. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon W B, Lin C, Palma J, Gao X Y, Wu S. J Biol Chem. 1993;268:5089–5096. [PubMed] [Google Scholar]
- 29.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S, Rowan S, Bani M R, Ben-David Y. Proc Natl Acad Sci USA. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amrolia P J, Ramamurthy L, Saluja D, Tanese N, Jane S M, Cunningham J M. Proc Natl Acad Sci USA. 1997;94:10051–10056. doi: 10.1073/pnas.94.19.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bean T L, Ney P A. Nucleic Acids Res. 1997;25:2509–2515. doi: 10.1093/nar/25.12.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotkow K J, Orkin S H. Mol Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai T, Igarashi K, Akasaka J, Furuyama K, Fujita H, Hayashi N, Yamamoto M, Sassa S. J Biol Chem. 1998;273:5358–5365. doi: 10.1074/jbc.273.9.5358. [DOI] [PubMed] [Google Scholar]
- 35.Racke F K, Lewandowska K, Goueli S, Goldfarb A N. J Biol Chem. 1997;272:23366–23370. doi: 10.1074/jbc.272.37.23366. [DOI] [PubMed] [Google Scholar]
- 36.Devemy E, Billat C, Haye B. Cell Signal. 1997;9:41–46. doi: 10.1016/s0898-6568(96)00095-2. [DOI] [PubMed] [Google Scholar]
- 37.Bittorf T, Jaster R, Ludtke B, Kamper B, Brock J. Cell Signal. 1997;9:85–89. doi: 10.1016/s0898-6568(96)00121-0. [DOI] [PubMed] [Google Scholar]
- 38.Schonwasser D C, Marais R M, Marshall C J, Parker P J. Mol Cell Biol. 1998;18:790–798. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crossley M, Orkin S H. J Biol Chem. 1994;269:16589–16596. [PubMed] [Google Scholar]
- 40.Cheng X, Reginato M J, Andrews N C, Lazar M A. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Yang X, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 43.Forsberg W C, Lam L T, Yang X-J, Nakatani Y, Bresnick E H. Biochemistry. 1997;36:15918–15924. doi: 10.1021/bi971664x. [DOI] [PubMed] [Google Scholar]
- 44.Gavva N R, Gavva R, Ermekova K, Sudol M, Shen C-K J. J Biol Chem. 1997;272:24105–24108. doi: 10.1074/jbc.272.39.24105. [DOI] [PubMed] [Google Scholar]
- 45.Imhof M O, McDonnell D P. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huibregtse J M, Scheffner S, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatakeyama S, Jensen J P, Weissman A M. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- 48.Karin M, Liu Z, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 49.Shivdasani R A, Rosenblatt M F, Zucker-Franklin D, Jackson C W, Hunt P, Saris C J M, Orkin S H. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 50.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 51.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]