Abstract
TcRζ/CD3 ligation initiates a signaling cascade involving CD4/CD8-p56lck, p59fyn, and ZAP-70, as well as lymphoid downstream proteins VAV, SLP-76, and FYB/SLAP. A current question concerns the nature of the downstream binding partner(s) of FYB in T cells. In this study, using a two-hybrid screen with FYB as bait, we have identified eight clones, four of which correspond to the recently published lymphoid protein SKAP55, and two which correspond to a related protein with some 44% homology to SKAP55 (termed SKAP55-related protein, SKAP55R). The SKAP55 clones showed only minor differences (two substitutions and one residue deletion) from SKAP55. SKAP55R has the same overall structure as SKAP55 except for the presence of a unique N terminus with a well-defined coiled-coil domain. Both SKAP55 and SKAP55R were found to bind FYB through their SH3 domains and to act as substrates for the FYN kinase in T cells. Furthermore, immunofluorescence confocal microscopy showed that FYB and SKAP55 colocalize in the perinuclear region of cells. SKAP55 also colocalizes with another FYB binding protein, SLP-76. Taken together, these observations demonstrate that FYB is part of an interactive matrix with SKAP55 and a SKAP55-related protein.
Ligation of CD4/CD8-p56lck and the T cell receptor complex (TcRζ/CD3) activates src protein-tyrosine kinases (PTKs) p56lck and p59fyn (1), leading to phosphorylation of immunoreceptor tyrosine-based activation motifs (ITAMs) of the TcRζ and CD3 chains (2–4). The phosphorylated ITAM recruits ZAP-70 by means of tandem SH2 domain binding (5, 6). CD4–p56lck can further up-regulate ZAP-70 catalytic activity by phosphorylation of residue Y-493 (7). The importance of ZAP-70 in T cell signaling has been shown by defects in early signaling events and interleukin 2 (IL-2) transcription in ZAP-70-negative Jurkat cells (8). The Lck-SH2 domain can also bind to ZAP-70, thereby consolidating the CD4/CD8-p56lck and TcRζ/CD3 aggregate (9). Consistent with this capability, p56lck has been found associated with the TcRζ/CD3 complex (10). p56lck may also play roles in signaling by other systems by virtue of its ability to phosphorylate other surface receptors such as CD5 and CD28 (11–13).
Recent advances have identified downstream targets of the TcR/CD3 and CD4/CD8-p56lck complexes that include hematopoietic proteins VAV, SLP-76, and FYB. VAV possesses several domains, including a guanine nucleotide exchange factor (GEF) domain for the Rho and Rac small GTP-binding proteins (14–16). In turn, VAV binds to another hematopoietic protein, SLP-76 (17–19), which possesses a C-terminal SH2 domain and proline-rich motifs (20). ZAP-70 is the key kinase that phosphorylates SLP-76 (21, 22), allowing the SH2 domain to bind to two pYESP motifs (Y-113 and Y-128) within the protein (22). VAV and SLP-76 cooperate to augment TcRζ/CD3 induced IL-2 transcription (17, 18). VAV–SLP-76 complex formation, however, is not essential for TcR-induced IL-2 production in all T cells (22). Instead, SLP-76 requires an intact SLP-76 SH2 domain in its potentiation of IL-2 transcription (18).
In this regard, we recently reported the cDNA encoding of a human and murine 120-kDa protein (termed FYB for FYN binding protein) that binds to the SH2 domains of FYN and SLP-76 (23–25). A similar protein, SLAP (for SLP-associated protein), has been cloned by others (26). Expression of FYB/SLAP is restricted to T cells, thymocytes, and myeloid cells and does not occur in B cells (25). It has several proline-rich sequences, multiple tyrosine-based motifs, two stretches of highly charged residues similar to nuclear localization sequences, and an SH3-like domain (25, 26). FYB/SLAP shows some basal phosphorylation in resting T cells, but it undergoes increased phosphorylation in response to TcR ligation (24). Consistent with the finding that FYB preferentially associates with FYN, T cells from FYN-negative mice show a marked reduction in FYB phosphorylation (24). FYB/SLAP has been implicated in IL-2 secretion (25) and in the negative regulation of IL-2 transcription (26). These observations point to a role for FYB in signaling mediated by SLP-76 and the FYN kinase. In this context, it is interesting that another lymphoid-specific protein, termed SKAP55 (src kinase-associated protein, 55 kDa), has been cloned and binds to FYN (27).
An important question in understanding the nature of this signaling cascade has been whether FYB/SLAP is linked to a downstream binding partner. In this report, using the two-hybrid system with FYB as bait, we have identified two FYB-binding proteins that correspond to SKAP55, and a SKAP55-related protein (termed SKAP55R). SKAP55R has the same overall structure as SKAP55 except for the presence of a unique N terminus with a coiled-coil domain. Both SKAP55 and SKAP55R bind FYB through their SH3 domains, and they act as substrates for the FYN kinase. Immunofluorescence confocal microscopy showed that FYB and SKAP55 colocalize in the perinuclear region of cells. Taken together, these observations demonstrate that FYB is part of an interactive matrix with SKAP55 and/or a SKAP55-related protein.
EXPERIMENTAL PROCEDURES
Cells and Antibodies.
The murine T cell hybridoma DC27.10 and COS cells were cultured in RPMI medium 1640 supplemented with 5% (vol/vol) fetal bovine serum (FBS; Intergen, Purchase, NY), 100 units/ml penicillin, 100 μg/ml streptomycin (GIBCO), 2 mM l-glutamine (GIBCO), and 50 μM 2-mercaptoethanol. The phenotype and function of the DC27.10 T cells have been previously described (28). Cells were transfected with FYN, FYB, SLP-76, or SKAP55 in the pSRα2 expression vector (gift of M. Streuli, Dana–Farber Cancer Institute). mAb to SLP-76 was kindly provided by Paul R. Findell (Syntex, Palo Alto, CA); GST-GRB-2 was kindly provided by Steve Shoelson (Joslin Diabetes Center, Boston). Anti-glutathione S-transferase (GST) was purchased from Santa Cruz Biotechnology, and anti-FYN mAb was from Transduction Laboratories (Lexington, KY). For electroporation studies, DC27.10 cells were subjected to 260 V and 960 μF in the presence of various DNA constructs.
Two-Hybrid Screen.
The DNA fragments encoding the full-length and C-terminal portion (amino acids 318–783) of human FYB protein were generated by PCR and inserted into the yeast two-hybrid vector pAS2–1 (CLONTECH) at the sites SfiI/SalI in frame fused to GAL4 binding domain. The recombinant plasmids were introduced into the yeast strain Y190 according to the procedure from the supplier. The yeast two-hybrid library from human peripheral blood leukocytes (Matchmaker Library) was purchased from CLONTECH (kind gift of Prasad Kanteti, Dana–Farber Cancer Institute). The library DNA was introduced into the yeast strain expressing the FYB fusion proteins. Positive clones were detected by β-galactosidase assay. Plasmid DNA was purified from positive clones, and both strands were subjected to sequencing by automatic sequencer (Dana–Farber Cancer Institute Molecular Core Facility). DNA sequences were analyzed by using the Genetics Computer Group (GCG) program (Madison, WI).
Immunoprecipitation and Immunoblotting.
Immunoprecipitation analysis and immunoblotting were conducted as previously described (28).
Immunofluorescence.
COS cells were cotransfected with FYB-120 and SKAP55 or SLP-76 and SKAP55 as described above. Cells were plated on glass coverslips and grown for 1 day prior to staining. Cells were rinsed with PBS, fixed with 2% paraformaldehyde in PBS for 10 min, and permeabilized for 10 min in 0.5% Triton X-100 in PBS. To detect FYB, SLP-76, and SKAP55, permeabilized cells were exposed to the specific antibodies in blocking buffer (2% normal goat serum and 10% rabbit serum in PBS) for 1 hr, followed by a brief wash and incubation with isotype-specific secondary antibody [labeled with Texas red or fluorescein isothiocyanate (FITC); Southern Biotechnology Associates] and 0.5 μg/ml Hoechst dye 33238 [Sigma; to visualize DNA (blue)]. Slides were mounted in a poly(vinyl alcohol) medium and immunofluorescence was analyzed with the Zeiss LSM410 confocal laser scanning microscope. The images were printed with the Fuji Pictrography 3000 color printer by using Adobe PhotoShop software (Adobe Systems, Mountain View, CA).
RESULTS
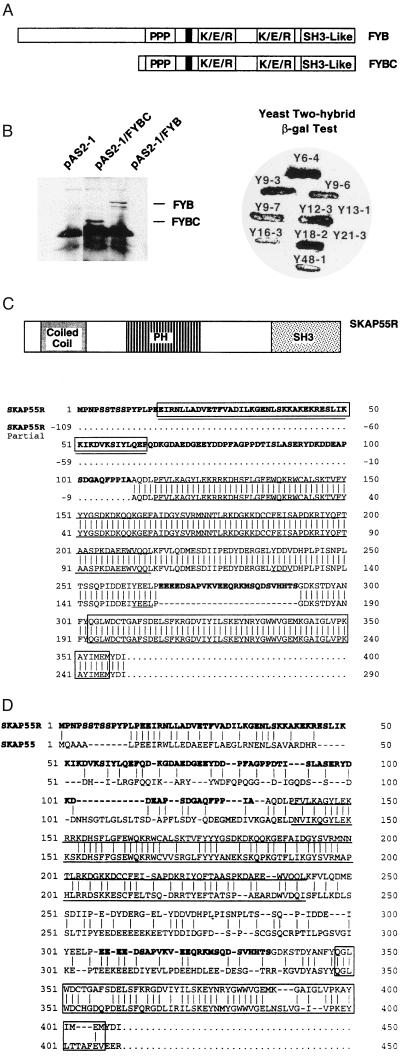
In an attempt to identify binding partners for FYB, the yeast two-hybrid system was used with FYB as bait, together with a cDNA library from peripheral blood lymphocytes. Bait was cloned into the yeast two-hybrid cloning vector pAS2–1 and used to transform the yeast strain Y190 (see Experimental Procedures). FYB contains several putative SH3-binding proline-rich sequences, multiple tyrosine-based motifs, two putative nuclear localization sequences, and a SH3-like domain (25). Two forms of FYB were used, a full-length and a truncated form (residue 317 to the C terminus) (Fig. 1A). The truncated version contained the two stretches of proline residues as well as the SH3-like domain. Full-length and truncated FYB were fused in-frame with the GAL4 binding domain and were initially examined for expression. Western blotting using polyclonal anti-FYB serum identified the expression of both forms at approximately 120 kDa and 80 kDa, respectively (Fig. 1B Left). These two clones were then used as bait to screen for FYB-binding proteins in a yeast two-hybrid cDNA library prepared from human peripheral blood lymphocytes. From this method, eight positive isolates from screening were found by the β-galactosidase assay (Fig. 1B Right). Plasmid DNA was prepared from these clones and subjected to DNA sequencing. All clones were found to bind to both the full-length and truncated forms of FYB in secondary screens.
Figure 1.
SKAP55 and SKAP55R clones identified by the two-hybrid screen using FYB as bait. (A) Depiction of full-length FYB and truncated FYB (FYBC) used in the two-hybrid screen. (B) (Left) Expression of FYB and FYBC in yeast. (Right) β-Galactosidase test using full-length FYB identified positive clones Y6-4. Y9-3, Y9-6, Y9-7, Y12-3, Y16-3, Y18-2, and Y48-1. Clones Y13-1 and Y21-3 served as negative controls. (C) Deduced amino acid sequence of the full-length SKAP55R protein. The amino acid sequence of the newly cloned N terminus and midregion insertion is shown in boldface. The PH domain is underlined in boldface, the putative src-SH2 binding motif is underlined, the coiled-coil region is designated by a combination of rectangular box and underline, and the SH3 domain is designated in a rectangular box. (D) Amino acid comparison of SKAP55 and full-length SKAP55R. The amino acid sequence of the newly cloned N terminus and midregion insertion is shown in boldface (upper line). The PH domain of both proteins is underlined in boldface and the SH3 domains are designated in a rectangular box.
Of the eight clones, four corresponded to the recently reported lymphoid protein SKAP55 (27). Only two or three residues were found to differ. In each of our clones, a serine was substituted for a threonine at residue 187 and a valine was absent at position 293 (data not shown). Interestingly, an additional difference was found among our individual clones at residue 161. Clones Y16-3, Y12-3, and Y9-3 had a serine at this site, whereas clone Y48-1 possessed a threonine. Because the library is derived from multiple donors, this finding suggested that this site acts to identify allelism in the SKAP55 gene.
In addition to the SKAP55 clones, clone Y18-2 carried a 2104-bp cDNA insert encoding a full-length sequence for a SKAP55-related protein (SKAP55R) with a calculated molecular mass of 41 kDa (Fig. 1C). A comparison of the predicted amino acid sequence with the database showed homology with a partial clone recently submitted to GenBank (submitted by R. Waterston, Washington Univ.; GenBank accession no. AC003999). The major difference was the presence in our full-length clone of an extended N-terminal sequence of 110 amino acids and a 26 amino acid insertion at residue 156. As does SKAP55, SKAP55R possesses a pleckstrin homology domain (PH domain; residues 115–214), multiple tyrosine phosphorylation sites, and a SH3 domain (residues 303–307) (Fig. 1C). While SKAP55R and SKAP55 showed 44% identity, the greatest overall conservation is found in the PH and in the SH3 domains (Fig. 1D). The tyrosine motif YEVL in SKAP55 exists as YEEL in SKAP55R, both of which could potentially serve as src kinase SH2 domain binding sites (29). Adjacent to this motif in SKAP55R exists a region of charged residues (i.e., EEEED; residues 266–290). The N terminus in the full-length SKAP55R has multiple residues that could serve as phosphorylation sites. These include YPLP (residues 11–14), YLQE (residues 59–62), YDDP (residues 75–79), and YDKD (residues 93–97). However, the major structural difference between SKAP55 and SKAP55R was found in the N-terminal region, where SKAP55R showed the presence of a well-defined coiled-coil (residues 20–75) (Fig. 1C). The coiled-coil is not present in SKAP55. Coiled-coils are formed by the coiling of two right-handed helices around each other with a slight left-handed superhelical twist (30, 31). They display a pattern of hydrophilic and hydrophobic residues that are repeated every seven residues (32).
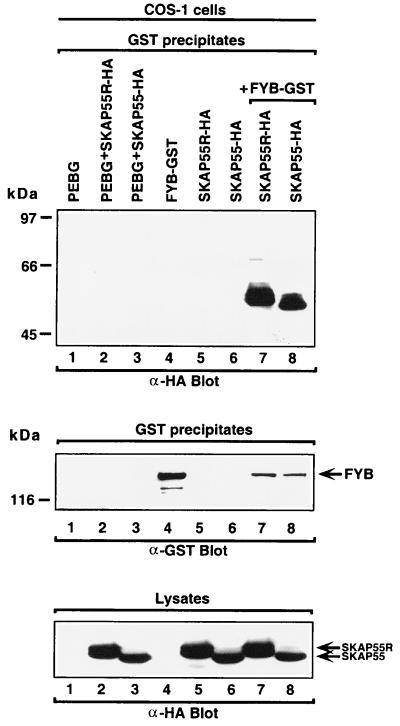
To verify SKAP55 and SKAP55R binding to FYB in vivo, the proteins were coexpressed in COS cells and assessed for binding. FYB was tagged with GST, whereas SKAP55 and SKAP55R were tagged with a hemagglutinin (HA)-derived peptide. As seen in Fig. 2, SKAP55R and SKAP55 were readily coprecipitated by FYB, as detected by anti-HA immunoblotting (Top, lanes 7, 8, respectively). SKAP55R was generally found to migrate slightly slower than SKAP55, possibly related to the presence of the N-terminal coiled-coil loop (lane 7 vs. 8). Roughly equivalent amounts of SKAP55 and SKAP55R were precipitated by FYB. In the absence of FYB, neither of the SKAP55 proteins was precipitated (lanes 2, 3, 5, and 6). The expression of 120-kDa FYB was monitored in cell lysates by anti-GST blotting (Fig. 2 Middle, lanes 4, 7, and 8). Similarly, SKAP55 and SKAP55R expression was measured in cell lysates by anti-HA blotting (Fig. 2 Bottom, lanes 2, 3, and 5–8). These data provide in vivo confirmation that full-length SKAP55 and SKAP55R are capable of binding to FYB.
Figure 2.
SKAP55 and SKAP55R bind to FYB in vivo. (Top) SKAP55 and SKAP55R bind to FYB in COS cells. COS cells were transfected with combinations of FYB, SKAP55, and SKAP55R and assessed for complex formation. Glutathione beads were used to precipitate the GST-fusion proteins, and anti-HA and anti-GST blotting followed. Lane 1, pEBG; lane 2, pEBG and SKAP55R-HA; lane 3, pEBG and SKAP55-HA; lane 4, FYB-GST; lane 5, SKAP55R-HA; lane 6, SKAP55-HA; lane 7, SKAP55R-HA and FYB-GST; and lane 8, SKAP55-HA and FYB-GST. (Middle) Levels of FYB protein expression. As in Top except GST precipitates were blotted with anti-GST. (Bottom) Levels of SKAP55 and SKAP55R expression. Cell lysates were blotted with anti-HA.
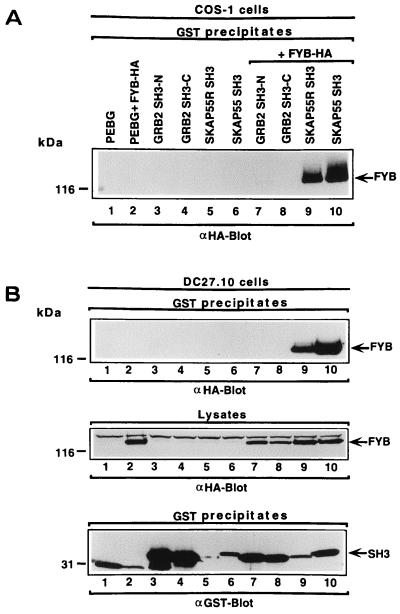
Key residues conserved in the SH3 domains are conserved in SKAP55 and SKAP55R (33). In the case of SKAP55R, these include a lysine at residue 305 of the βa region, a conserved tryptophan doublet within the βc region at residues 336–337, a proline at residue 349 of the βd–βe region, and a tyrosine at residue 352 of the βe region. The SH3 domains of SKAP55 and SKAP55R are highly similar except in the βd–βc region, where some six residues differ. Other key residues such as the tryptophan doublet within the βc region, the proline in the βd–βe region, and the tyrosine in the βe region, are conserved. Given this similarity, we next assessed whether the SH3 domains of SKAP55 and SKAP55R could bind to FYB. To test this possibility, GST-SH3 domains of the individual proteins were coexpressed with HA-tagged FYB in COS cells or in the T cell hybridoma DC27.10. Under these conditions, the SKAP55R and SKAP55 SH3 domains were found to precipitate FYB (Fig. 3A, lanes 9 and 10, respectively). As a control for specificity, neither the N- nor C-terminal Grb-2 SH3 domain was found to interact with FYB (lanes 7 and 8). The same GRB-2 preparations had been previously shown to precipitate other proteins such as Son of Sevenless and CD28 (34). Anti-HA blotting demonstrated equivalent levels of FYB expression in transfected cells, and, similarly, the various fusion proteins were well expressed, as detected by anti-GST blotting (data not shown).
Figure 3.
SKAP55 and SKAP55R SH3 domains bind to FYB. (A) SKAP55 and SKAP55R SH3 domains bind to FYB in COS cells. COS cells were transfected with combinations of FYB, SKAP55, SKAP55R, and GRB-2 SH3 domains and assessed for complex formation. SH3 domain-containing proteins were expressed as GST fusion proteins. Glutathione beads were used to precipitate the GST-fusion proteins and anti-HA blotting followed. Lane 1, pEBG; lane 2, pEBG and FYB-HA; lane 3, Grb-2 N-terminal SH3 domain; lane 4, Grb-2 C-terminal SH3 domain; lane 5, SKAP55R SH3 domain; lane 6, SKAP55 SH3 domain; lane 7, Grb-2 N-terminal SH3 domain and FYB-HA; lane 8, Grb-2 C-terminal SH3 domain and FYB-HA; lane 9, SKAP55R SH3 domain and FYB-HA; lane 10, SKAP55 SH3 domain and FYB-HA. (B) (Top) SKAP55 and SKAP55R SH3 domains bind to FYB in T cells. DC27.10 cells were transfected with FYB and SKAP55, SKAP55R, and GRB-2 SH3 domains and assessed for complex formation. Lanes as in A. (Middle) Levels of FYB protein expression. As in Top except cell lysate was blotted with anti-HA. (Bottom) Levels of GST-fusion protein expression. As in Top except anti-GST blot.
It was next important to demonstrate that this interaction could occur in T cells. The T cell hybridoma DC27.10 has been used in previous function and signaling studies (35, 36). When the approach described above was used, both the SKAP55R and SKAP55 SH3 domains precipitated FYB from transfected DC27.10 T cells (Fig. 3B Top, lanes 9 and 10, respectively), whereas neither Grb-2 SH3 domain was found to interact with the protein (lanes 7 and 8). Anti-HA and anti-GST blotting confirmed expression of FYB and GST proteins in cells (Fig. 3 Middle and Bottom, respectively). These data demonstrate that SKAP55 and SKAP55R can bind in vivo to FYB through their SH3 domains in T cells.
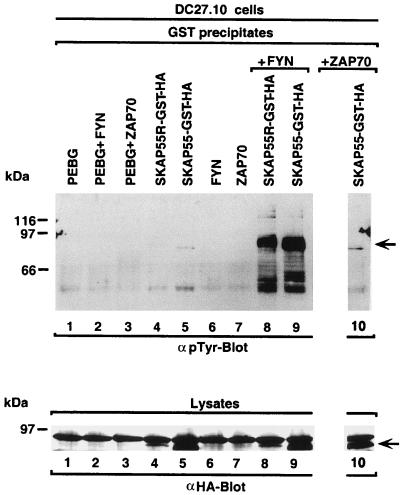
Coincident with our finding that SKAP55 and FYB bind to each other is a previous observation that these two proteins bind selectively to the src kinase FYN (25, 27). We have previously shown that FYN can phosphorylate FYB (25). However, the kinase responsible for the phosphorylation of SKAP55 has not been defined. Our observation of binding between SKAP55 and FYB further supported the possibility that the FYN kinase might phosphorylate both proteins. To examine this question, SKAP55 tagged with GST and HA was coexpressed with FYN in the T cell hybridoma, precipitated from cell lysates, and assessed for tyrosine phosphorylation by anti-phosphotyrosine blotting (Fig. 4). Under these conditions, SKAP55 showed a marked phosphorylation by FYN (Fig. 4 Upper, lane 9) (the band with lower molecular mass correspond to partially degraded forms of SKAP55). In the absence of the kinase, SKAP55 failed to show any phosphorylation (lane 5). As a negative control, expression of another T cell kinase, ZAP-70, failed to phosphorylate the protein (lane 10). Previous studies using the same reagents have shown the presence of ZAP-70 activity in its preferential phosphorylation of another lymphoid protein, SLP-76 (22). As a control for expression, blotting of lysates from cells transfected with HA-tagged proteins showed consistent levels of SKAP55 expression (Fig. 4 Lower, lanes 5, 9, and 10). FYN and ZAP-70 expression was also found to be consistent between samples (data not shown). SKAP55R also shared an ability to act as a substrate for the FYN kinase (Fig. 4 Upper, lane 8). Again, without kinase, no SKAP55R phosphorylation was observed (lane 4). Blotting of lysates from transfected cells showed levels of SKAP55R expression that were lower than SKAP55, but nevertheless sufficient to allow for detectable phosphorylation by FYN (Fig. 4 Upper, lanes 4 vs. 8). These data indicate that in addition to binding to FYB, SKAP55 and SKAP55R share an ability to act as substrates for the FYN kinase.
Figure 4.
SKAP55 and SKAP55R act as targets of FYN kinase. (Upper) SKAP55 and SKAP55R act as substrates for FYN. DC27.10 cells were transfected with SKAP55 or SKAP55R and FYN or ZAP-70 and assessed for tyrosine phosphorylation. SKAP55 and SKAP55R were tagged with both GST and HA. GST precipitates were subjected to anti-phosphotyrosine blotting (lanes 7–10). Lane 1, pEBG; lane 2, pEBG and FYN; lane 3, pEBG and ZAP-70; lane 4, SKAP55R-GST-HA; lane 5, SKAP55-GST-HA; lane 6, FYN; lane 7, ZAP-70; lane 8, SKAP55R-GST-HA plus FYN; lane 9, SKAP55-GST-HA plus FYN; lane 10, SKAP55-GST-HA plus ZAP-70. (Lower) Expression levels of SKAP55-GST-HA and SKAP55R-GST-HA. As in Upper except cell lysates were blotted with anti-HA.
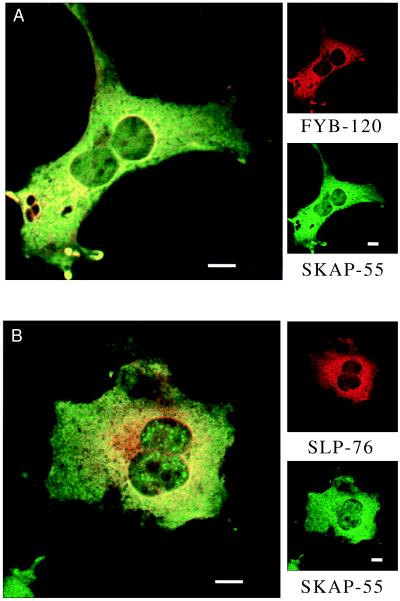
Given the binding between SKAP55 and FYB, it was of interest to assess whether these proteins colocalize in cells. Confocal immunofluorescence microscopy was used to visualize SKAP55 and FYB in transfected COS cells with FITC-and Texas red-labeled secondary antibodies. Previous work on lymphoid proteins such as Lck noted staining primarily at cell surface (ref. 37 and data not shown). By contrast, FYB and SKAP55 were detected mostly in the cytoplasm, in particular, around the nucleus (Fig. 5A, Upper and Lower Right, respectively). Only a small amount of plasma membrane staining was occasionally evident. Further, an overlay of FYB and SKAP55 staining (double stained for FYB (Texas red, red) and SKAP55 (FITC, green) revealed colocalization as seen with visible yellow fluorescence indicative of coincident staining (Fig. 5A Left). FYB perinuclear staining was confirmed by setting of the threshold of intensity where the greatest levels were found in this region (data not shown). SKAP55 tended to show a slightly broader pattern of staining in the cytoplasm compared with FYB. Similar levels of colocalization were observed in T cells, although a detailed visualization of intracellular localization was not possible given the small amount of cytoplasm (data not shown). These observations are consistent with FYB and SKAP55 binding in cells.
Figure 5.
Co-localization of FYB with SKAP55 and SLP76 with SKAP55 as detected by confocal immunofluorescence. COS cells transfected with FYB-120, SLP-76, and/or SKAP55 were stained with a combination of FITC- and Texas red-labeled secondary antibodies. (A) FYB and SKAP55 show intracellular colocalization. (Upper Right) Anti-FYB-120. (Lower Right) Anti-SKAP55. (Left) Anti-FYB and anti-SKAP55. (B) SLP-76 and SKAP55 show intracellular colocalization. (Upper Right) Anti-SLP-76. (Lower Right) Anti-SKAP55. (Left) Anti-SKAP55 and anti-SLP-76. (Scale bars correspond to 10 μm.)
Because we and others previously showed that FYB/SLAP could also interact with SLP-76 (25, 26), we were also interested in assessing whether SLP-76 and SKAP55 could colocalize. From Fig. 5B it is evident that SLP-76 and SKAP55 also colocalized in the intracellular compartment, around the nucleus. Furthermore, occasionally, SKAP55 also showed some nuclear staining with the appearance of concentric dot-like arrays in the nucleus. These observations support the idea that SKAP55 and SLP-76, in binding to FYB, colocalize in the cytoplasm.
DISCUSSION
T cell activation involves the activation of p56lck, p59fyn-T, and ZAP-70 and the phosphorylation of an array of downstream targets that include lymphoid-specific proteins VAV, SLP-76, FYB/SLAP, and SKAP55. A connection between these proteins first became evident with the demonstration of VAV binding to SLP-76 (17–19), ZAP-70 phosphorylation of SLP-76 (21, 22), and regulation by ZAP-70 of VAV-SLP-76 binding (22). More recently, FYB/SLAP-130 has been identified as a selective target of the FYN and SLP-76 SH2 domains (24, 26). Each of these observations points to the presence of an interactive matrix of lymphoid proteins in T cells. A major question was whether another component interacts with FYB in the signaling cascade. In this study, we demonstrate by use of the two-hybrid system and by in vivo coexpression studies that FYB binds to two proteins, SKAP55 and SKAP55R. Of eight positive clones, four corresponded to SKAP55 and three to SKAP55R. Our data identified the full-length sequence for a SKAP55-related protein that shares 44% identity with SKAP55 and retains functional domains such as the PH domain for membrane localization, a putative src kinase SH2 binding motif [YEEL (residues 261–264)], and a SH3 domain. However, SKAP55R differed from SKAP55 in the presence of a well defined coiled-coil domain in the N-terminal region. Coiled-coil domains tend to undergo tight binding to each other and are found in fibrous proteins such as tropomyosin, myosin, and keratins, as well as transcriptional activators such as the homeobox-containing protein Ubx, the zinc finger protein Rpt1, and the prokaryotic transcriptional activator FixJ (38). SKAP55R might therefore be expected to generate signals similar to those of SKAP55, as well as unique signals, or have a unique localization due to the N-terminal region. Further work will be needed to compare the properties of SKAP55 and SKAP55R. Given their size and sequence similarity, SKAP55 and SKAP55R likely originated as a result of gene duplication. The identification of SKAP55 and SKAP55R binding to FYB extends the complexity of proteins known to bind FYB, and it offers the possibility that SKAP55 and SKAP55R will act downstream in a TcR signaling cascade. The perinuclear localization and the occasional appearance of SKAP55 in the nucleus support this scenario.
Consistent with their conserved nature, the SH3 domains of SKAP55 and SKAP55R were found to mediate binding to FYB. This mediation was demonstrated in vivo by coexpression with FYB in COS and T cells (Fig. 3). The two domains are conserved in key residues such as the tryptophan doublet within the βc region, the proline in the βd-βe region, and the tyrosine in the βe region. No obvious difference was observed in the efficiency of FYB precipitation by the two SH3 domains. By contrast, the GRB-2 C- and N-terminal SH3 domains failed to precipitate FYB, pointing to the specificity of the SKAP55/SKAP55R SH3 domain interaction. Although the binding sites on FYB have not been mapped, the protein is highly enriched with proline residues, including PXXP motifs that could serve as sites of binding (24, 26).
FYN phosphorylation of SKAP55 and SKAP55R provided further evidence in support of a connection between these proteins and FYB. SKAP55 and SKAP55R shared an ability to serve as targets for FYN, whereas another kinase, ZAP-70, failed to induce significant phosphorylation (Fig. 4). SKAP55, SKAP55R, and FYB could therefore coexist in a matrix influenced by the FYN kinase. This observation fits with findings that FYB and SKAP55 bind to the SH2 domain of FYN (25, 27). FYB and SKAP-55 also showed a unique intracellular colocalization that differs from the restricted localization of p56lck at the plasma membrane (37) and provides an explanation for the absence of FYB or SKAP55 in anti-Lck precipitates (25, 27). Colocalization of SKAP55 with the other FYB binding protein, SLP-76, further supports the idea of a lymphoid-specific matrix mediated by FYB binding. SLP-76 binds to FYB by means of its SH2 domain (25). No direct binding between SLP-76 and SKAP55 has been observed (data not shown). FYB could possibly act as a scaffold in binding to SKAP55, SKAP55R, FYN, and SLP-76. Once bound, these proteins could potentially interact and influence each other’s function.
FYB and SKAP55 binding to and phosphorylation by FYN suggest that they may be connected to T cell signaling by means of a connection with this src kinase. Functions attributed to FYN such as the regulation of TcR-mediated IL-2 production (39) and the induction of anergy (40, 41) might therefore be mediated downstream by FYB and SKAP55. Future studies will be needed to assess the role of FYB, SKAP55, and SKAP55R in T cell function.
Acknowledgments
C.E.R is the recipient of a Scholar Award from the Leukemia Society of America. This work was supported by National Institutes of Health Grant CA51887.
ABBREVIATIONS
- TcR
T cell receptor
- IL-2
interleukin 2
- FYB
FYN binding protein
- SKAP55
src kinase-associated protein of 55 kDa
- SKAP55R
SKAP55-related protein
- GST
glutathione S-transferase
- HA
hemagglutinin
- FITC
fluorescein isothiocyanate
Note Added in Proof
A comparison of sequences determined by our laboratory and that of Dr. B. Schraven (Heidelberg) has shown that our SKAP55-R sequence is essentially the same (one residue difference) as the sequence for another cloned variant of SKAP55 termed SKAP55 HOM (A. Marie-Cardine and B. Schraven, personal communication).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF072166).
References
- 1.Rudd C E, Janssen O, Cai Y-C, da Silva A J, Raab M, Prasad K V S. Immunol Today. 1994;15:225–234. doi: 10.1016/0167-5699(94)90248-8. [DOI] [PubMed] [Google Scholar]
- 2.Weiss A, Littman D R. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 3.Mustelin T. Immunity. 1994;1:351–356. doi: 10.1016/1074-7613(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 4.Wange R L, Samelson L E. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- 5.Chan A C, Iwashima M, Turck C W, Weiss A. Cell. 1992;71:649–662. doi: 10.1016/0092-8674(92)90598-7. [DOI] [PubMed] [Google Scholar]
- 6.Iwashima M, Irving B A, Van Oers N S C, Chan A C, Weiss A. Science. 1994;263:1136–1139. [PubMed] [Google Scholar]
- 7.Chan A C, Dalton M, Johnson R, Kong G H, Wang T, Thoma R, Kurosaki T. EMBO J. 1995;14:2499–2508. doi: 10.1002/j.1460-2075.1995.tb07247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams B L, Schreiber K L, Zhang W, Wange R, Samelson L, Leibson P J, Abraham R T. Mol Cell Biol. 1998;18:1388–1399. doi: 10.1128/mcb.18.3.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duplay P, Thome M, Herve F, Acuto O. J Exp Med. 1994;179:1163–1172. doi: 10.1084/jem.179.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess K E, Odysseos A D, Zalvan C, Druker B J, Anderson P, Schlossman S F, Rudd C E. Eur J Immunol. 1991;21:1663–1668. doi: 10.1002/eji.1830210712. [DOI] [PubMed] [Google Scholar]
- 11.Burgess E K, Yamamoto M, Prasad K V S, Rudd C E. Proc Natl Acad Sci USA. 1992;89:9311–9315. doi: 10.1073/pnas.89.19.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raab M, Cai Y-C, Bunnell S C, Heyeck S D, Berg L J, Rudd C E. Proc Natl Acad Sci USA. 1995;92:8891–8895. doi: 10.1073/pnas.92.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raab M, Yamamoto M, Rudd C E. Mol Cell Biol. 1994;14:2862–2870. doi: 10.1128/mcb.14.5.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch C A, Anderson D, Moran M F. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 15.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo X R. Nature (London) 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 16.Collins T L, Deckert M, Altman A. Immunol Today. 1997;18:221–225. doi: 10.1016/s0167-5699(97)01037-2. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Motto D G, Koretsky G A, Weiss A. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- 18.Motto D G, Ross S E, Wu J, Hendriks-Taylor L R, Koretzky G A. J Exp Med. 1996;183:1937–1943. doi: 10.1084/jem.183.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onodera H, Motto D G, Koretzky G A, Rothstein D M. J Biol Chem. 1996;271:22225–22230. doi: 10.1074/jbc.271.36.22225. [DOI] [PubMed] [Google Scholar]
- 20.Jackman J K, Motto D G, Sun Q, Tanemoto M, Turck C W, Peltz G A, Koretzky G A, Findell P R. J Biol Chem. 1995;270:7029–7032. doi: 10.1074/jbc.270.13.7029. [DOI] [PubMed] [Google Scholar]
- 21.Wardenburg J B, Fu C, Jackman J K, Flotow H, Wilkinson S E, Williams D H, Johnson R, Kong G, Chan A C, Findell P R. J Biol Chem. 1996;271:19641–19644. doi: 10.1074/jbc.271.33.19641. [DOI] [PubMed] [Google Scholar]
- 22.Raab M, da Silva A J, Findell P R, Rudd C E. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80422-7. [DOI] [PubMed] [Google Scholar]
- 23.da Silva A J, Rudd C E. J Biol Chem. 1993;268:16537–16543. [PubMed] [Google Scholar]
- 24.da Silva A J, Rosenfield J M, Mueller I, Bouton A, Hirai H, Rudd C E. J Immunol. 1997;158:2007–2016. [PubMed] [Google Scholar]
- 25.da Silva A J, Li Z, de Vera C, Canto E, Findell P, Rudd C E. Proc Natl Acad Sci USA. 1997;94:7493–7498. doi: 10.1073/pnas.94.14.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musci M A, Hendricks-Taylor L R, Motto D G, Paskind M, Kamens J, Turck C W, Koretzky G A. J Biol Chem. 1997;272:11674–11677. doi: 10.1074/jbc.272.18.11674. [DOI] [PubMed] [Google Scholar]
- 27.Marie-Cardine A, Bruyns E, Eckerskorn C, Kirchgessner H, Meuer S, Schraven B. J Biol Chem. 1997;272:16077–16080. doi: 10.1074/jbc.272.26.16077. [DOI] [PubMed] [Google Scholar]
- 28.Cai Y-C, Cefai D, Schneider H, Raab M, Nabavi N, Rudd C E. Immunity. 1995;3:1–10. doi: 10.1016/1074-7613(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 29.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 30.Crick F H C. Acta Crystallogr. 1953;6:689. [Google Scholar]
- 31.Pauling L, Corey R B. Nature (London) 1953;171:59–61. doi: 10.1038/171059a0. [DOI] [PubMed] [Google Scholar]
- 32.Cohen C, Parey D A D. Trends Biochem Sci. 1986;11:245–247. [Google Scholar]
- 33.Mayer B J, Eck M J. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 34.Kim H H, Tharaygil M, Rudd C E. J Biol Chem. 1997;273:296–301. doi: 10.1074/jbc.273.1.296. [DOI] [PubMed] [Google Scholar]
- 35.Zamoyska R, Derham P, Gorman S D, von Hoegen P, Bolen J B, Veillette A, Parnes J R. Nature (London) 1989;342:278–281. doi: 10.1038/342278a0. [DOI] [PubMed] [Google Scholar]
- 36.Cai Y-C, Cefai D, Schneider H, Raab M, Nabavi N, Rudd C E. Immunity. 1995;3:417–426. doi: 10.1016/1074-7613(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 37.Ley S C, Marsh M, Bebbington C R, Proudfoot K, Jordan P. J Cell Biol. 1994;125:639–649. doi: 10.1083/jcb.125.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 39.Davidson D, Chow L, Fournel M, Veillette A. J Exp Med. 1992;175:1483–1492. doi: 10.1084/jem.175.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quill H, Riley M P, Cho E A, Casnellie J E, Reed J, Torigoe T. J Immunol. 1992;149:2887–2893. [PubMed] [Google Scholar]
- 41.Boussiotis V A, Barber D L, Lee B J, Gribben J G, Freeman G J, Nadler L M. J Exp Med. 1996;184:365–376. doi: 10.1084/jem.184.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]