Abstract
During the maturation of the immune response, antibody genes are subjected to localized hypermutation. Mutations are not evenly distributed along the V gene; intrinsic hot spots exist that are correlated with primary sequence motifs. Although the mechanism of hypermutation remains unknown, it has been proposed to exhibit DNA strand polarity because purine residues on the coding strand are more frequently targeted for mutation than pyrimidines. However, this polarity may not be an intrinsic property of the hypermutation mechanism but a consequence of evolutionary-selected peculiarities of V gene sequences. Furthermore, the possibility that both strands are hypermutation targets has received little attention. To discriminate between these possibilities, we have analyzed the average frequency of mutations of each of the three bases of all nucleotide triplets by using large databases taken from both V and non-V mutation targets. We also have reassessed the sequence motifs associated with hot spots. We find that even in non-Ig sequences, A mutates more than T, consistent with a strand-dependent component to targeting. However, the mutation biases of triplets and of their inverted complements are correlated, demonstrating that there is a sequence-specific but strand-independent component to mutational targeting. Thus, there are two aspects of the hypermutation process that are sensitive to local DNA sequences, one that is DNA strand-dependent and the other that is not.
During the maturation of the immune response, antibody genes hypermutate. This process, highly specific for the immune system, is characterized by the introduction of point mutations at a very high rate. It occurs only within a DNA segment of ≈1–2 Kb, encompassing the bulk of the V region but excluding the C. The B cells expressing the somatically mutated variants are then subjected to an antigen-mediated selection resulting in affinity maturation (reviewed in refs. 1 and 2).
The frequency at which the four bases hypermutate suggests a strand bias. In particular, in the transcribed strand, T residues accumulate fewer mutations than A despite the fact that they are a complementary pair (3–5). This point has been used to suggest that the mutations occur on only one DNA strand and is consistent with many hypermutation models (3, 4, 6–9). However, it remains possible that the observed strand discrimination is caused, at least in part, by the nonrandom nature of hypermutation. The nonrandom distribution of intrinsic mutations is highlighted by hot as well as cold spots. There is formal proof that short sequence motifs are associated with hot spots (10, 11), but other interactions additionally have been postulated to account for the diverse mutability of the same motif when found in different DNA segments (10, 12, 13) Thus, the nonrandom, sequence-dependent distribution of hot spots also could give rise to strand discrimination.
It is not readily feasible to formally establish in vivo whether hypermutation targets only one or both DNA strands, but the problem can be approached indirectly because the rate of mutation of each base depends on its local environment. In the case of Ig V genes, this environment is unlikely to be random. Indeed, analysis of codon usage in Ig V genes strongly indicates that their DNA sequences have evolved to ensure strategic localization of somatic hypermutation hot spots (14). However, by analysis of mutation in V gene flanking sequences or in transgenic non-Ig targets (11, 15), the pattern of nucleotide substitutions can be examined in sequences that are unlikely to have been subjected to evolutionary selection for nonrandom distribution of hot spots. Here, by using large databases of such mutations, we contrast the mutation distributions observed with what would have been anticipated if either one or both DNA strands are hypermutation targets.
MATERIALS AND METHODS
Strategy of the Analysis.
We analyzed the coding strand to establish the degree of correlation between the average mutation frequency of individual bases of triplets and of their inverted complements. Significant correlation is to be expected if both strands are hypermutation substrates. Thus, if both strands are targeted equally, the mutability of a given triplet on the coding strand should equal that of its inverted complement (e.g., 5′-TAC and 5′-GTA, respectively).
Obviously, the reliability of our estimates of the mutation frequencies in each data set depends on the number of mutated sequences analyzed. Within each data set, these ranged from 37 to 224 (Table 1), which we assume are sufficient for meaningful conclusions. Pooling all data into a single database would have given undue weight to the sets represented by the largest number of sequences. Thus, we separately calculated the mean mutation frequency for each base type in every triplet of our data sets, and only then were the values pooled.
Table 1.
Mutation databases
Computation and Statistical Analysis.
Let S be the number of sequences in each of the sets analyzed (Table 1). All triplets are counted so that each overlaps its nearest neighbors by two bases. Let wijk be the number of occurrences of a given triplet (i, j, k = T, A, C, or G) in each wild-type sequence and mijkp the number of mutations in position p (p = first, second, or third base of triplet). The percentage mutation frequency of bases in the triplets of each set was
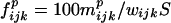 |
and their mean frequency (F) for the n sets (typically seven)
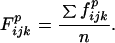 |
The coefficient of correlation (r) was calculated by using MGLH regression routines of the program systat (Ver. 5.2, SYSTAT Inc., Evanston, IL)
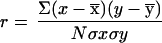 |
where x and y are mutation frequencies of base x and its complementary base y, respectively, x̄ and ȳ are their mean values, and σxσy are the corresponding SDs. n = 32 (triplet pairs) for correlation between mutations of the second base of triplets and their corresponding inverted complements, and n = 64 for correlation between mutations of the first and third base of the inverted complements. A check of the calculations was performed by randomly scrambling the mutation frequencies to produce 1 million new sets of pairs and recalculating the correlation coefficients.
RESULTS
Distribution of Mutations: Nucleotide Preferences and Hotspots.
Mutations were analyzed in seven target sequences. Three (VκOx1Jκ5, Vjλ1, and VH26) code for V segments of mouse light and human heavy chains, one spans the noncoding 3′-flank of mouse VHDJH rearrangement (JH4 flank), and the other three (gpt, neo, and globin) are heterologous non-Ig DNA sequences that have been inserted into transgenes as artificial mutation targets (Table 1).
It has been suggested that hypermutation exhibits strand polarity because, in the coding strand, A was found more mutated than T and sometimes G more than C (3, 4, 12, 15). The analysis of our database confirmed a consistent imbalance only in the frequency at which A and T are mutated (Table 2).
Table 2.
Preferential targeting of A residues
| Relative mutation*
|
||||
|---|---|---|---|---|
| T | C | A | G | |
| VκOx1Jκ5 | 0.14 | 0.23 | 0.32 | 0.31 |
| VH26 | 0.20 | 0.18 | 0.31 | 0.31 |
| VJλ1 + 3′intron | 0.17 | 0.21 | 0.36 | 0.26 |
| JH4 | 0.23 | 0.21 | 0.38 | 0.18 |
| gpt | 0.21 | 0.31 | 0.25 | 0.23 |
| neo | 0.22 | 0.17 | 0.40 | 0.21 |
| Globin | 0.23 | 0.29 | 0.27 | 0.21 |
| Average | 0.20 | 0.23 | 0.33 | 0.24 |
Corrected for base composition.
With regard to individual triplets, they are present in differing numbers in the various data sets. Some are present only once or are even absent in a given data set (e.g., TTT, TTG, and TAG in VkOx1; see also Table 3). With such low numbers, differences of mutation averages in individual data sets are very unreliable. This problem clearly illustrates the importance of multiple data sets for meaningful calculations. Indeed, leaving aside the poorly represented CpG containing triplets, the mutation frequencies of the pooled data derived from 22 (e.g., TAA) to 77 (e.g., TGG) independent mutated triplets.
Table 3.
Examples of variability of triplet frequencies and of mutation frequency of their middle base
| Triplet | gpt | JH | Globin |
|---|---|---|---|
| CTA | 6.2/2 | 1.6/8 | 4.1/3 |
| AAC | 2.2/3 | 5.2/3 | 0.9/3 |
| ATA | 4.5/5 | 3.4/3 | 0/0 |
| GCT | 7.3/3 | 2.2/8 | 5.6/3 |
| TAT | 3.6/6 | 5.3/9 | 4.1/1 |
| AGC | 6.7/4 | 5.0/7 | 8.8/2 |
Relative mutation frequency (%) of the middle base/number of times the triplet occurs in the gene fragment.
Analyzing the compiled data (Tables 4 and 5) for evidence of consensus hot spots revealed that triplets containing highly mutated bases were often related to known hot spot motifs. However, not all of the bases within such triplets were highly mutated. For instance, the bracketed residues (A)GC, GC(T), and TA(C) were much less mutated than the other two within each triplet. The highest scoring triplets (3.0 or above) were aligned to derive an independent consensus of a longer motif (Table 6). The alignment could be rationalized assuming that there are two types of motifs, namely G-A-G/a-C/t-T/a (lower case indicating lower mutation frequency) and T-A-T/C/G. In addition, the inverted complements were often also high scorers. If both strands of DNA are hypermutation targets (as we argue below), a single base could have been targeted preferentially, the palindromic neighbor reflecting to some extent the targeting of a similar hot spot motif in the second strand. Thus, the hot spot motifs may be more accurately described for the coding strand as the consensus of Table 6, where the underlined bases are the main primary target. These hot spot motifs differ somewhat from those previously proposed (16). It may be relevant that biochemical analysis of replication fidelity by pure polymerase α showed considerable misincorporation at the base in italics in the sequences TTACG, TTGCA, and AAGCT (17). These sequences have considerable similarity with the Ig hot spot motifs.
Table 4.
Correlation between frequency of mutation of the middle base of all triplets and their inverted repeats
| Triplet | Wx/Wy | F2x/F2y |
|---|---|---|
| TTT/AAA | 64/40 | 1.5/1.9 |
| TTC/GAA | 43/32 | 0.9/1.7 |
| TTA/TAA | 29/22 | 2.1/2.4 |
| TTG/CAA | 51/43 | 0.8/1.0 |
| TCT/AGA | 71/44 | 1.1/1.0 |
| TCC/GGA | 46/52 | 0.9/0.5 |
| TCA/TGA | 53/52 | 0.3/0.7 |
| TCG/CGA | 13/12 | 0.3/0.6 |
| TAT/ATA | 41/25 | 5.0/4.3 |
| TAC/GTA | 30/31 | 4.2/2.1 |
| TAG/CTA | 28/36 | 4.6/3.0 |
| TGT/ACA | 59/40 | 1.7/0.9 |
| TGC/GCA | 50/50 | 2.9/2.0 |
| TGG/CCA | 77/48 | 0.6/0.4 |
| CTT/AAG | 43/43 | 1.9/3.1 |
| CTC/GAG | 62/52 | 0.7/1.1 |
| CTG/CAG | 84/69 | 0.7/0.9 |
| CCT/AGG | 47/60 | 0.9/0.6 |
| CCC/GGG | 25/56 | 0.8/0.3 |
| CCG/CGG | 19/13 | 0.1/0.4 |
| CAT/ATG | 35/44 | 1.8/1.0 |
| CAC/GTG | 45/59 | 1.6/0.9 |
| CGT/ACG | 10/8 | 2.2/1.2 |
| CGC/GCG | 17/12 | 1.6/0.4 |
| ATT/AAT | 42/27 | 1.8/3.1 |
| ATC/GAT | 34/43 | 0.7/1.6 |
| ACT/AGT | 47/48 | 2.2/3.2 |
| ACC/GGT | 39/53 | 1.6/2.6 |
| AAC/GTT | 28/37 | 4.1/1.7 |
| AGC/GCT | 39/59 | 5.8/4.1 |
| GTC/GAC | 43/31 | 0.6/1.1 |
| GCC/GGC | 31/45 | 2.0/2.7 |
The database involves all fragments of Table 1. The first column shows the triplets compared. The second shows the total respective number computed. The third depicts the ratio of the mutation frequency of their middle base. For other details, see Materials and Methods.
Table 5.
Correlation between frequency of mutation of the first base of all triplets and of the third base of their inverted repeats
| Triplet | Wx/Wy | F1x/F3y |
|---|---|---|
| TTT/AAA | 64/40 | 1.5/2.5 |
| TTC/GAA | 43/32 | 3.0/3.9 |
| TTA/TAA | 29/22 | 1.1/1.6 |
| TTG/CAA | 51/43 | 1.2/2.8 |
| TCT/AGA | 71/44 | 0.5/1.0 |
| TCC/GGA | 46/52 | 1.1/1.4 |
| TCA/TGA | 53/52 | 0.8/1.9 |
| TCG/CGA | 13/12 | 0.9/1.4 |
| TAT/ATA | 41/25 | 1.8/3.3 |
| TAC/GTA | 30/31 | 4.0/6.8 |
| TAA/TTA | 22/29 | 2.0/2.5 |
| TAG/CTA | 28/36 | 3.0/3.2 |
| TGT/ACA | 59/40 | 0.8/0.8 |
| TGC/GCA | 50/50 | 0.9/1.6 |
| TGA/TCA | 52/53 | 0.6/1.4 |
| TGG/CCA | 77/48 | 1.0/1.5 |
| CTT/AAG | 43/43 | 3.1/1.4 |
| CTC/GAG | 62/52 | 2.3/5.7 |
| CTA/TAG | 36/28 | 3.7/3.3 |
| CTG/CAG | 84/69 | 1.6/2.3 |
| CCT/AGG | 47/60 | 1.0/1.5 |
| CCC/GGG | 25/56 | 1.9/2.2 |
| CCA/TGG | 48/77 | 1.8/1.7 |
| CCG/CGG | 19/13 | 1.3/1.4 |
| CAT/ATG | 35/44 | 0.9/1.6 |
| CAC/GTG | 45/59 | 0.6/1.8 |
| CAA/TTG | 43/51 | 0.7/0.9 |
| CAG/CTG | 69/84 | 1.3/1.2 |
| CGT/ACG | 10/8 | 0.1/2.8 |
| CGC/GCG | 17/12 | 0.9/1.3 |
| CGA/TCG | 12/13 | 1.3/0.3 |
| CGG/CCG | 13/19 | 0.0/1.1 |
| ATT/AAT | 42/27 | 2.8/1.5 |
| ATC/GAT | 34/43 | 3.1/2.7 |
| ATA/TAT | 25/41 | 3.5/1.0 |
| ATG/CAT | 44/35 | 2.9/2.3 |
| ACT/AGT | 47/48 | 2.6/1.2 |
| ACC/GGT | 39/53 | 2.8/1.4 |
| ACA/TGT | 40/59 | 2.7/1.4 |
| ACG/CGT | 8/10 | 1.2/1.3 |
| AAT/ATT | 27/42 | 1.6/1.6 |
| AAC/GTT | 28/37 | 2.5/0.9 |
| AAA/TTT | 40/64 | 1.0/1.2 |
| AAG/CTT | 43/43 | 1.5/1.4 |
| AGT/ACT | 48/47 | 1.8/1.1 |
| AGC/GCT | 39/59 | 1.2/1.1 |
| AGA/TCT | 44/71 | 1.7/1.4 |
| AGG/CCT | 60/47 | 2.1/0.9 |
| GTT/AAC | 37/28 | 3.3/2.9 |
| GTC/GAC | 43/31 | 0.7/0.9 |
| GTA/TAC | 31/30 | 5.0/1.8 |
| GTG/CAC | 59/45 | 1.4/1.2 |
| GCT/AGC | 59/39 | 6.0/5.3 |
| GCC/GGC | 31/45 | 0.7/0.5 |
| GCA/TGC | 50/50 | 3.3/3.5 |
| GCG/CGC | 12/17 | 1.5/1.2 |
| GAT/ATC | 43/34 | 0.8/1.6 |
| GAC/GTC | 31/43 | 0.4/0.3 |
| GAA/TTC | 32/43 | 0.7/0.6 |
| GAG/CTC | 52/62 | 0.5/0.5 |
| GGT/ACC | 53/39 | 0.6/1.3 |
| GGC/GCC | 45/31 | 0.3/0.1 |
| GGA/TCC | 52/46 | 0.9/0.7 |
| GGG/CCC | 56/25 | 0.2/0.3 |
The database involves all fragments of Table 1. The first column shows the pair to be compared. The second shows the respective number of computed triplets. The third depicts the ratio of the mutation frequency of the first base of each triplet over the third of its inverted complement. For other details, see Materials and Methods.
Table 6.
Hot spot consensus sequences of the coding strand
| AGC, | 5.8 | (GCT, 4.1) |
| GCT, | 6.0 | (AGC, 5.3) |
| TAC, | 4.2 | |
| GAG, | 5.7 | |
| GTA, | 5.0 | |
| GAA, | 3.9 | (TTC, 3.0) |
| AAC, | 4.1 | |
| AGT, | 3.2 | |
| AAT, | 3.1 | |
| AAG, | 3.1 | |
| GCA, | 3.3 | (TGC, 3.5) |
| GTT, | 3.3 | |
| TAG, | 3.0 | (CTA, 3.7) |
| Consensus | ||
| G-A-G/a-C/t-T/A | ||
| TAT, | 5.0 | (ATA, 4.3) |
| GTA, | 6.8 | (TAC, 4.0) |
| ATA, | 3.3 | |
| TAG, | 4.6 | (CTA 3.0) |
| CTA, | 3.2 | (TAG, 3.0) |
| TAC, | 4.2 | |
| ATA, | 3.5 | |
| ATC, | 3.1 | |
| Consensus | ||
| T-A-T/C/G/a. | ||
Numbers indicate mutation frequency of underlined base. In brackets are the corresponding inverted repeats.
Correlation of Mutation Frequency of Triplets and Their Inverted Complements.
The mutation frequency of individual bases of all triplets and their inverted complements (Tables 4 and 5) were found to be correlated, with a confidence level well above 99.9% (Table 7). Although the corresponding correlation coefficients were substantial, there were some instructive anomalies. For example, the mutation frequencies of the bases in italics of GAG and CTC were 5.7 and 2.3, respectively. The main reason for this discrepancy was traced to the skewing effect of the most prominent hot spot in the gpt data set (15). Indeed, although this site represented only 3% of all GAGs in the integrated database, it contributed with 24% of the GAG mutations (data not shown). Another major discrepancy was GTA/TAC (5.0/1.8). In this case, two of the GTA triplets of VH26 representing 4.7% of the total database accounted for 30% of G mutations. Those discrepancies thus originate from individual bases being exceptionally targeted for mutation owing to features of their sequence environment not included within the spanning nucleotide triplet.
Table 7.
Correlation of mutation frequency of individual bases of all triplets and of their inverted complements
| Data sets | Bases compared* | r† | P‡ |
|---|---|---|---|
| All sets | 2 vs. 2 | 0.74 | 0.1 10−5 |
| All sets | 1 vs. 3 | 0.64 | 0.1 10−7 |
| Coding§ | 2 vs. 2 | 0.50 | 0.004 |
| Coding§ | 1 vs. 3 | 0.49 | 0.4 10−4 |
| Not coding§ | 2 vs. 2 | 0.64 | 0.7 10−4 |
| Not coding§ | 1 vs. 3 | 0.57 | 0.1 10−7 |
| W/palindromes | 2 vs. 2 | 0.56 | 0.016 |
| W/palindromes | 1 vs. 3 | 0.44 | 0.007 |
| No palindromes | 2 vs. 2 | 0.80 | 0.0006 |
| No palindromes | 1 vs. 3 | 0.74 | 0.8 10−5 |
| Control (all) | 3 vs. 3 | 0.08 | 0.68 |
Numbers indicate the position of the base in the triplets.
Correlation coefficient.
Probability of no correlation.
Coding or not coding for antibody.
Several controls were performed to check the validity of the analysis. For instance, no correlation in mutation frequency was found between the third bases of triplets and the third (as opposed to the first) base of their inverted complements (Table 7). Furthermore, the correlation remained statistically significant for separately computed groups of triplets, containing or excluding palindromic dinucleotides (Table 7).
The database also was divided into sequences that had or had not been subjected to antigenic selection. One fraction contained the k, λ, and heavy chain coding segments. The other contained the noncoding (3′ introns) of λ and heavy chains as well as sequences coding for irrelevant genes (Table 7). Relative to the total pool, the correlation was less marked in both fractions. Thus, the correlation increased with the size of the database rather than with selected subdivisions. Even so, the fraction comprising the non-Ig coding fraction showed a marginally better correlation than the Ig coding fraction, which could have reflected the evolutionary pressure to accumulate hot spots in antigen binding segments (14). However, other trivial explanations are also possible.
DISCUSSION
Both Strand-Independent and Strand-Dependent Components to Mutational Targeting.
The results of this paper reveal that there must be a component of mutational targeting that is sensitive to local sequence environment and that does not show strand discrimination. So much is indicated by the correlation between targeting frequencies of bases of individual triplets with that of their inverted complements. A paper published when this manuscript was completed reaches a similar conclusion analyzing the sequence motifs of hot spots (18).
However, in addition to this sequence-specific but strand-independent mutational targeting, evidence remains that the hypermutation process also exhibits some strand discrimination. It has long been suggested that the process of hypermutation displays strand polarity because A and G in the coding strand were found to be more mutated than T and C, respectively (3, 12, 15, 19) (not confirmed in ref. 18). Here, we also found consistent disparity between A and T (but not G and C) mutations. The size and complexity of our database makes it unlikely that the bias occurs by chance or as a consequence of evolutionary pressures because the non-Ig targets are unlikely to have been subjected to similar pressures. Furthermore, the results of Table 6 also point toward strand polarity in that the triplets representing the hot spot motifs are more mutated than their inverted complements. An element of the chain polarity is likely therefore to be due to preferential targeting of one of the two DNA strands. Thus, whereas the triplet analysis reveals that there is a sequence-dependent, strand-independent component to mutational targeting, the preferential targeting of A as opposed to T residues for mutation, argues in favor of an additional strand-dependent component to the targeting.
In conclusion, our analysis indicates that there may be two distinct phases of the hypermutation process that are sensitive to local DNA sequence environment. The two phases differ, however, in their strand discrimination.
Acknowledgments
We are grateful to Dr. C. Rada for her help in preparing the databases and for critically reading the manuscript. C.M. acknowledges the support of the National Foundation for Cancer Research.
References
- 1.Milstein C, Neuberger M S. Adv Prot Chem. 1996;49:451–485. doi: 10.1016/s0065-3233(08)60494-5. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 3.Lebecque S G, Gearhart P J. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuberger M S, Milstein C. Curr Opin Immunol. 1995;7:248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 5.Reynaud C A, Garcia C, Hein W R, Weill J C. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S, Milstein C. Nature (London) 1966;211:242–243. [Google Scholar]
- 7.Milstein C, Rada C. The Maturation of the Antibody Response. London: Academic; 1995. [Google Scholar]
- 8.Peters A, Storb U. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 9.Manser T. Immunol Today. 1990;11:305–308. doi: 10.1016/0167-5699(90)90124-r. [DOI] [PubMed] [Google Scholar]
- 10.Goyenechea B, Milstein C. Proc Natl Acad Sci USA. 1996;93:13979–13984. doi: 10.1073/pnas.93.24.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klix N, Jolly C J, Davies S L, Bruggemann M, Williams G T, Neuberger M S. Eur J Immunol. 1998;28:317–326. doi: 10.1002/(SICI)1521-4141(199801)28:01<317::AID-IMMU317>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 12.Betz A G, Rada C, Pannell R, Milstein C, Neuberger M S. Proc Natl Acad Sci USA. 1993;90:2385–2388. doi: 10.1073/pnas.90.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolly C J, Wagner S D, Rada C, Klix N, Milstein C, Neuberger M S. Semin Immunol. 1996;8:159–168. doi: 10.1006/smim.1996.0020. [DOI] [PubMed] [Google Scholar]
- 14.Wagner S D, Milstein C, Neuberger M S. Nature (London) 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- 15.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui Y L, Fernandez A G, Pannell R, Neuberger M S, Milstein C. Nature (London) 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 16.Rogozin I B, Sredneva N E, Kolchanov N A. Biochim Biophys Acta. 1996;1306:171–178. doi: 10.1016/0167-4781(95)00241-3. [DOI] [PubMed] [Google Scholar]
- 17.Goodman M F, Creighton S, Bloom L B, Petruska J. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 18.Dorner T, Foster S J, Brezinschek H-P, Lipsky P E. Immunol Rev. 1998;162:161–171. doi: 10.1111/j.1600-065x.1998.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 19.TumasBrundage K, Manser T. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez F A, Milstein C. Proc Natl Acad Sci USA. 1993;90:9862–9866. doi: 10.1073/pnas.90.21.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rada C, Gonzalez Fernandez A, Jarvis J M, Milstein C. Eur J Immunol. 1994;24:1453–1457. doi: 10.1002/eji.1830240632. [DOI] [PubMed] [Google Scholar]
- 22.Goyenechea B, Klix N, Yelamos J, Williams G T, Riddell A, Neuberger M S, Milstein C. EMBO J. 1997;16:3987–3994. doi: 10.1093/emboj/16.13.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner S D, Elvin J G, Norris P, Mcgregor J M, Neuberger M S. Int Immunol. 1996;8:701–705. doi: 10.1093/intimm/8.5.701. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez Fernandez A, Gupta S K, Pannell R, Neuberger M S, Milstein C. Proc Natl Acad Sci USA. 1994;91:12614–12618. doi: 10.1073/pnas.91.26.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]


