Abstract
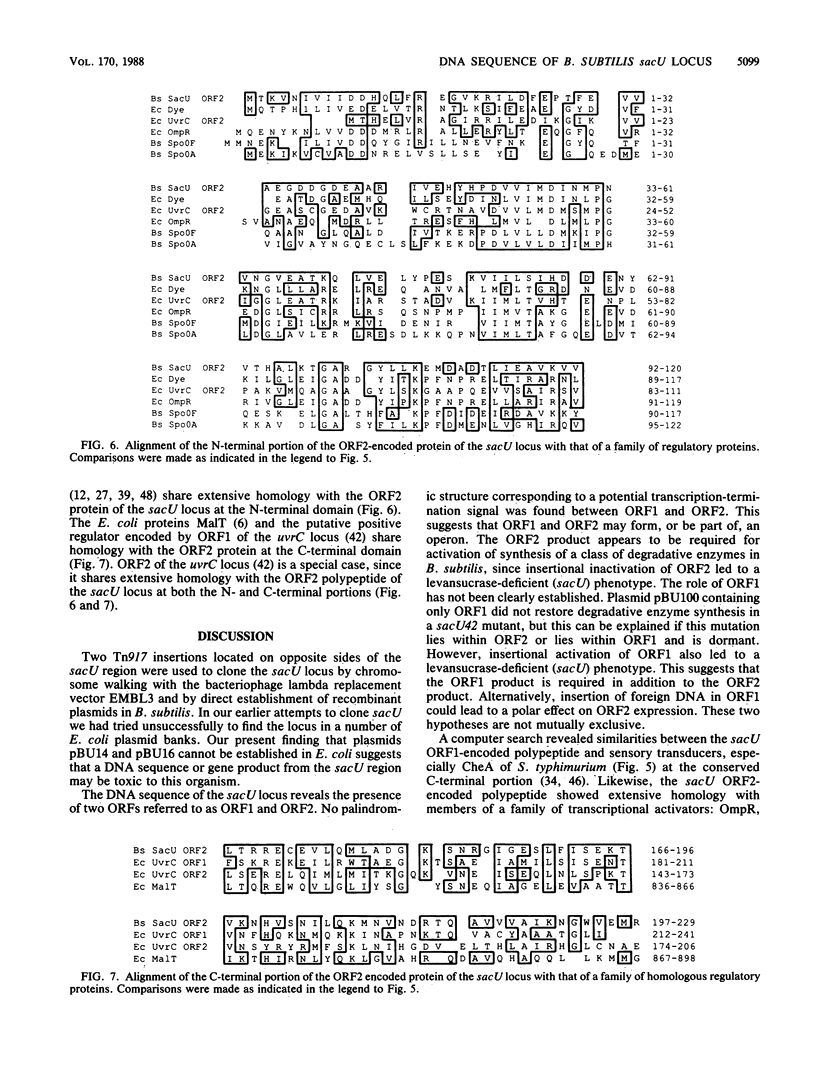
The sacU locus has been cloned by using two independent strategies, and the presence of two open reading frames was deduced from the nucleotide sequence. Open reading frame 1 encodes a 45,000-dalton polypeptide that is similar to the products of the Salmonella typhimurium cheA and Escherichia coli cpxA genes, which act as sensory transducers. Open reading frame 2 encodes a 26,000-dalton polypeptide that is similar to a family of transcriptional activators, including the products of the Bacillus subtilis spoOA and spoOF and the E. coli ompR and dye genes. These similarities suggest that the products of the B. subtilis sacU locus form a sensor-transducer couple, which functions to relay information about specific environmental changes to the transcription apparatus.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Hahn J., Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amory A., Kunst F., Aubert E., Klier A., Rapoport G. Characterization of the sacQ genes from Bacillus licheniformis and Bacillus subtilis. J Bacteriol. 1987 Jan;169(1):324–333. doi: 10.1128/jb.169.1.324-333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich S., Gonzy-Tréboul G., Steinmetz M. 5'-noncoding region sacR is the target of all identified regulation affecting the levansucrase gene in Bacillus subtilis. J Bacteriol. 1986 Jun;166(3):993–998. doi: 10.1128/jb.166.3.993-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayusawa D., Yoneda Y., Yamane K., Maruo B. Pleiotropic phenomena in autolytic enzyme(s) content, flagellation, and simultaneous hyperproduction of extracellular alpha-amylase and protease in a Bacillus subtilis mutant. J Bacteriol. 1975 Oct;124(1):459–469. doi: 10.1128/jb.124.1.459-469.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Raibaud O. The nucleotide sequence of the malT gene encoding the positive regulator of the Escherichia coli maltose regulon. Gene. 1986;42(2):201–208. doi: 10.1016/0378-1119(86)90297-0. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Drummond M., Whitty P., Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986 Feb;5(2):441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L. S., Buxton R. S. DNA sequence analysis of the dye gene of Escherichia coli reveals amino acid homology between the dye and OmpR proteins. J Biol Chem. 1985 Apr 10;260(7):4236–4242. [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Hoch J. A. Isolation of Bacillus subtilis genes from a charon 4A library. J Bacteriol. 1981 Apr;146(1):430–432. doi: 10.1128/jb.146.1.430-432.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Grossberger D. Minipreps of DNA from bacteriophage lambda. Nucleic Acids Res. 1987 Aug 25;15(16):6737–6737. doi: 10.1093/nar/15.16.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Albano M., Dubnau D. Isolation and characterization of Tn917lac-generated competence mutants of Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3104–3109. doi: 10.1128/jb.169.7.3104-3109.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Ferrari E., Perego M., Hoch J. A. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J Bacteriol. 1988 Jan;170(1):296–300. doi: 10.1128/jb.170.1.296-300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Yang M., Ferrari E. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signalling systems. J Bacteriol. 1988 Nov;170(11):5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aiba S. Isolation and characterization of antibiotic resistance plasmids from thermophilic bacilli and construction of deletion plasmids. J Bacteriol. 1981 Jun;146(3):1091–1097. doi: 10.1128/jb.146.3.1091-1097.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamata D., Gross J. D. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol Gen Genet. 1970;108(3):277–287. doi: 10.1007/BF00283358. [DOI] [PubMed] [Google Scholar]
- Klier A., Fouet A., Débarbouillé M., Kunst F., Rapoport G. Distinct control sites located upstream from the levansucrase gene of Bacillus subtilis. Mol Microbiol. 1987 Sep;1(2):233–241. doi: 10.1111/j.1365-2958.1987.tb00517.x. [DOI] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lepesant-Kejzlarova J., Lepesant J. A., Billault A., Dedonder R. Pleiotropic mutations affecting sporulation conditions and the syntheses of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56(11-12):1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- Leblanc D. J., Lee L. N. Physical and genetic analyses of streptococcal plasmid pAM beta 1 and cloning of its replication region. J Bacteriol. 1984 Feb;157(2):445–453. doi: 10.1128/jb.157.2.445-453.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant J. A., Kunst F., Lepesant-Kejzlarová J., Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol Gen Genet. 1972;118(2):135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- Lewandoski M., Dubnau E., Smith I. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J Bacteriol. 1986 Nov;168(2):870–877. doi: 10.1128/jb.168.2.870-877.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Mizuno T., Mizushima S. Interaction between two regulatory proteins in osmoregulatory expression of ompF and ompC genes in Escherichia coli: a novel ompR mutation suppresses pleiotropic defects caused by an envZ mutation. J Bacteriol. 1986 Dec;168(3):1309–1314. doi: 10.1128/jb.168.3.1309-1314.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh N., Simon M. I. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986 Jan;165(1):161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagami Y., Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Ehrlich S. D. In vitro genetic labeling of Bacillus subtilis cryptic plasmid pHV400. Plasmid. 1979 Jan;2(1):48–58. doi: 10.1016/0147-619x(79)90005-2. [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa K., Hess J. F., Simon M. I. Mutants defective in bacterial chemotaxis show modified protein phosphorylation. Cell. 1988 Apr 8;53(1):89–96. doi: 10.1016/0092-8674(88)90490-4. [DOI] [PubMed] [Google Scholar]
- Ostroff G. R., Pène J. J. Molecular cloning with bifunctional plasmid vectors in Bacillus subtilis: isolation of a spontaneous mutant of Bacillus subtilis with enhanced transformability for Escherichia coli-propagated chimeric plasmid DNA. J Bacteriol. 1983 Nov;156(2):934–936. doi: 10.1128/jb.156.2.934-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. B., Youngman P. J. Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Jan;83(1):140–144. doi: 10.1073/pnas.83.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooley H. M., Paschoud D., Karamata D. The gtaB marker in Bacillus subtilis 168 is associated with a deficiency in UDPglucose pyrophosphorylase. J Gen Microbiol. 1987 Dec;133(12):3481–3493. doi: 10.1099/00221287-133-12-3481. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Ruppen M. E., Van Alstine G. L., Band L. Control of intracellular serine protease expression in Bacillus subtilis. J Bacteriol. 1988 Jan;170(1):136–140. doi: 10.1128/jb.170.1.136-140.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Stark T. F., Beattie W. G., Moses R. E. Multiple control elements for the uvrC gene unit of Escherichia coli. Nucleic Acids Res. 1986 Mar 11;14(5):2301–2318. doi: 10.1093/nar/14.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes. J Bacteriol. 1986 Oct;168(1):380–388. doi: 10.1128/jb.168.1.380-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Kunst F., Dedonder R. Mapping of mutations affecting synthesis of exocellular enzymes in Bacillus subtilis. Identity of the sacUh, amyB and pap mutations. Mol Gen Genet. 1976 Nov 17;148(3):281–285. doi: 10.1007/BF00332902. [DOI] [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawata M. Cloning and characterization of Bacillus subtilis iep, which has positive and negative effects on production of extracellular proteases. J Bacteriol. 1988 Aug;170(8):3593–3600. doi: 10.1128/jb.170.8.3593-3600.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawata M., Nagami Y., Uchiyama H. prtR enhances the mRNA level of the Bacillus subtilis extracellular proteases. J Bacteriol. 1987 Jul;169(7):3044–3050. doi: 10.1128/jb.169.7.3044-3050.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Wylie D., Stock A., Wong C. Y., Stock J. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem Biophys Res Commun. 1988 Mar 15;151(2):891–896. doi: 10.1016/s0006-291x(88)80365-6. [DOI] [PubMed] [Google Scholar]
- Yang M., Ferrari E., Chen E., Henner D. J. Identification of the pleiotropic sacQ gene of Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):113–119. doi: 10.1128/jb.166.1.113-119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Shimotsu H., Ferrari E., Henner D. J. Characterization and mapping of the Bacillus subtilis prtR gene. J Bacteriol. 1987 Jan;169(1):434–437. doi: 10.1128/jb.169.1.434-437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Perkins J. B., Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195(3):424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]