Abstract
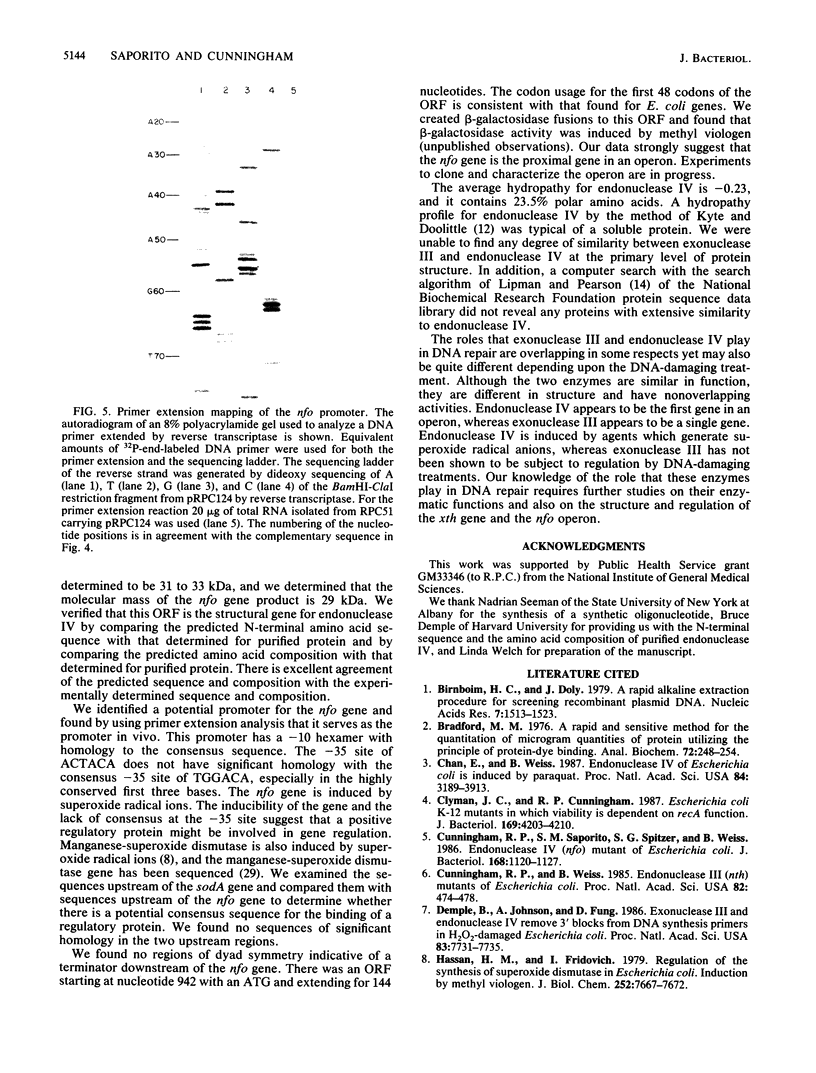
The nfo gene of Escherichia coli K-12 which encodes endonuclease IV has been sequenced. The predicted gene product has a molecular weight of 31,562, in good agreement with the size of the gene product estimated by maxicell analysis. The nfo promoter was mapped by primer extension of in vivo transcripts. Inspection of the nucleotide sequence revealed no regions of potential secondary structure corresponding to a transcriptional terminator downstream from the structural gene; however, there was a potential open reading frame immediately downstream from the nfo structural gene.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chan E., Weiss B. Endonuclease IV of Escherichia coli is induced by paraquat. Proc Natl Acad Sci U S A. 1987 May;84(10):3189–3193. doi: 10.1073/pnas.84.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyman J., Cunningham R. P. Escherichia coli K-12 mutants in which viability is dependent on recA function. J Bacteriol. 1987 Sep;169(9):4203–4210. doi: 10.1128/jb.169.9.4203-4210.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., Saporito S. M., Spitzer S. G., Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986 Dec;168(3):1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Johnson A., Fung D. Exonuclease III and endonuclease IV remove 3' blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H. M., Fridovich I. Regulation of the synthesis of superoxide dismutase in Escherichia coli. Induction by methyl viologen. J Biol Chem. 1977 Nov 10;252(21):7667–7672. [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner W. D., Grunberg S. M., Haseltine W. A. Enzyme action at 3' termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983 Dec 25;258(24):15198–15205. [PubMed] [Google Scholar]
- Kow Y. W., Wallace S. S. Exonuclease III recognizes urea residues in oxidized DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8354–8358. doi: 10.1073/pnas.82.24.8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Levin J. D., Johnson A. W., Demple B. Homogeneous Escherichia coli endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. J Biol Chem. 1988 Jun 15;263(17):8066–8071. [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J Biol Chem. 1977 May 10;252(9):2808–2814. [PubMed] [Google Scholar]
- Ljungquist S., Lindahl T., Howard-Flanders P. Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol. 1976 May;126(2):646–653. doi: 10.1128/jb.126.2.646-653.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3182–3186. doi: 10.1073/pnas.82.10.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Houlgrave C. W. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry. 1988 May 17;27(10):3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- Rogers S. G., Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65(1):201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. Cloning of uvrA, lexC and ssb genes of Escherichia coli. Biochem Biophys Res Commun. 1979 Sep 12;90(1):123–129. doi: 10.1016/0006-291x(79)91598-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporito S. M., Smith-White B. J., Cunningham R. P. Nucleotide sequence of the xth gene of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4542–4547. doi: 10.1128/jb.170.10.4542-4547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Avila H. Structure and gene expression of the E. coli Mn-superoxide dismutase gene. Nucleic Acids Res. 1986 Jun 11;14(11):4577–4589. doi: 10.1093/nar/14.11.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]