Abstract
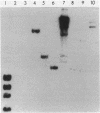
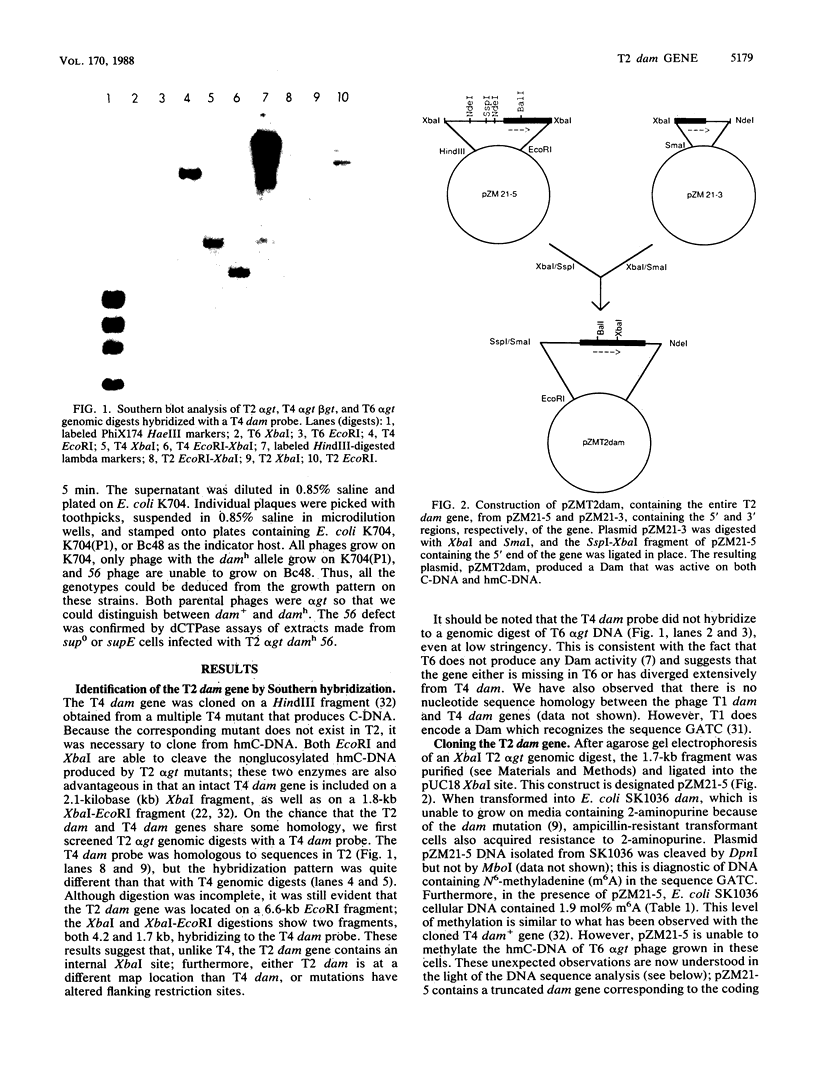
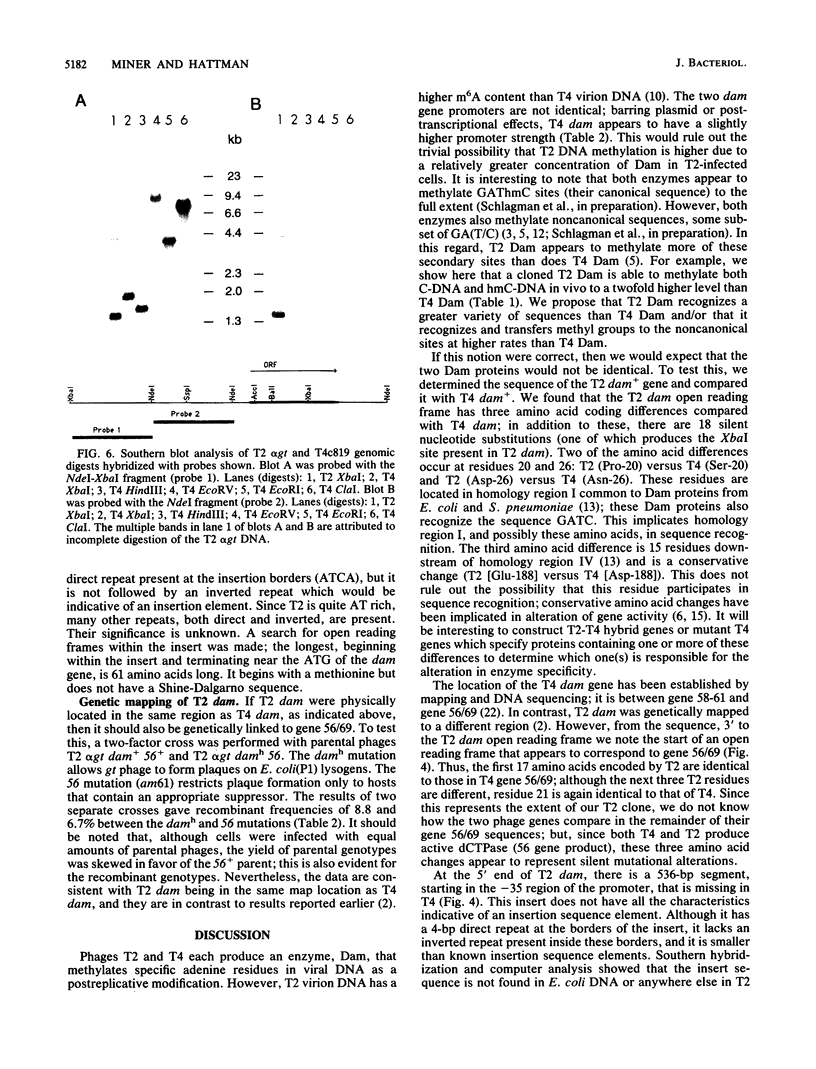
Bacteriophage T2 codes for a DNA-(adenine-N6)methyltransferase (Dam), which is able to methylate both cytosine- and hydroxymethylcytosine-containing DNAs to a greater extent than the corresponding methyltransferase encoded by bacteriophage T4. We have cloned and sequenced the T2 dam gene and compared it with the T4 dam gene. In the Dam coding region, there are 22 nucleotide differences, 4 of which result in three coding differences (2 are in the same codon). Two of the amino acid alterations are located in a region of homology that is shared by T2 and T4 Dam, Escherichia coli Dam, and the modification enzyme of Streptococcus pneumoniae, all of which methylate the sequence 5' GATC 3'. The T2 dam and T4 dam promoters are not identical and appear to have slightly different efficiencies; when fused to the E. coli lacZ gene, the T4 promoter produces about twofold more beta-galactosidase activity than does the T2 promoter. In our first attempt to isolate T2 dam, a truncated gene was cloned on a 1.67-kilobase XbaI fragment. This construct produces a chimeric protein composed of the first 163 amino acids of T2 Dam followed by 83 amino acids coded by the pUC18 vector. Surprisingly, the chimera has Dam activity, but only on cytosine-containing DNA. Genetic and physical analyses place the T2 dam gene at the same respective map location as the T4 dam gene. However, relative to T4, T2 contains an insertion of 536 base pairs 5' to the dam gene. Southern blot hybridization and computer analysis failed to reveal any homology between this insert and either T4 or E. coli DNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brooks J. E., Hattman S. In vitro methylation of bacteriophage lambda DNA by wild type (dam+) and mutant (damh) forms of the phage T2 DNA adenine methylase. J Mol Biol. 1978 Dec 15;126(3):381–394. doi: 10.1016/0022-2836(78)90047-5. [DOI] [PubMed] [Google Scholar]
- Brooks J., Hattman S. Location of the DNA-adenine methylase gene on the genetic map of phage T2. Virology. 1973 Sep;55(1):285–288. doi: 10.1016/s0042-6822(73)81032-3. [DOI] [PubMed] [Google Scholar]
- Doolittle M. M., Sirotkin K. Bacteriophage T2 and T4, dam+ and damh and Eco dam+ methylation: preference at different sites. Biochim Biophys Acta. 1988 Feb 28;949(2):240–246. doi: 10.1016/0167-4781(88)90088-7. [DOI] [PubMed] [Google Scholar]
- Douglas C. M., Collier R. J. Exotoxin A of Pseudomonas aeruginosa: substitution of glutamic acid 553 with aspartic acid drastically reduces toxicity and enzymatic activity. J Bacteriol. 1987 Nov;169(11):4967–4971. doi: 10.1128/jb.169.11.4967-4971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M., Hausmann R., Gold M., Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966 May 10;241(9):1995–2006. [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methylation of T-even bacteriophages and of their nonglucosylated mutants: its role in P1-directed restriction. Virology. 1970 Oct;42(2):359–367. doi: 10.1016/0042-6822(70)90279-5. [DOI] [PubMed] [Google Scholar]
- Hattman S., Wilkinson J., Swinton D., Schlagman S., Macdonald P. M., Mosig G. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J Bacteriol. 1985 Nov;164(2):932–937. doi: 10.1128/jb.164.2.932-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Hausmann R., Gold M. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. IX. Deoxyribonucleic acid methylase in bacteriophage-infected Escherichia coli. J Biol Chem. 1966 May 10;241(9):1985–1994. [PubMed] [Google Scholar]
- Heaphy S., Singh M., Gait M. J. Effect of single amino acid changes in the region of the adenylylation site of T4 RNA ligase. Biochemistry. 1987 Mar 24;26(6):1688–1696. doi: 10.1021/bi00380a030. [DOI] [PubMed] [Google Scholar]
- Hehlmann R., Hattman S. Mutants of bacteriophage T2 gt with altered DNA methylase activity. J Mol Biol. 1972 Jun 28;67(3):351–360. doi: 10.1016/0022-2836(72)90455-x. [DOI] [PubMed] [Google Scholar]
- Heitman J., Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987 Jul;169(7):3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli K-12 clones that overproduce dam methylase are hypermutable. J Bacteriol. 1981 Jan;145(1):644–646. doi: 10.1128/jb.145.1.644-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- Lauster R., Kriebardis A., Guschlbauer W. The GATATC-modification enzyme EcoRV is closely related to the GATC-recognizing methyltransferases DpnII and dam from E. coli and phage T4. FEBS Lett. 1987 Aug 10;220(1):167–176. doi: 10.1016/0014-5793(87)80897-9. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Mosig G. Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J. 1984 Dec 1;3(12):2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Mannarelli B. M., Balganesh T. S., Greenberg B., Springhorn S. S., Lacks S. A. Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4468–4472. doi: 10.1073/pnas.82.13.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton N. P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984 Nov;31(1-3):269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- Revel H. R. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967 Apr;31(4):688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer E., Auer B., Schweiger M. Identification, purification, and characterization of Escherichia coli virus T1 DNA methyltransferase. J Biol Chem. 1987 Nov 5;262(31):15225–15231. [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S. Molecular cloning of a functional dam+ gene coding for phage T4 DNA adenine methylase. Gene. 1983 May-Jun;22(2-3):139–156. doi: 10.1016/0378-1119(83)90098-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yee J. K., Marsh R. C. Alignment of a restriction map with the genetic map of bacteriophage T4. J Virol. 1981 Apr;38(1):115–124. doi: 10.1128/jvi.38.1.115-124.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ormondt H., Gorter J., Havelaar K. J., de Waard A. Specificity of deoxyribonucleic acid transmethylase induced by bacteriophage T2. I. Nucleotide sequences isolated from tmicrococcus luteus DNA methylated in vitro. Nucleic Acids Res. 1975 Aug;2(8):1391–1400. doi: 10.1093/nar/2.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]