Abstract
The von Hippel–Lindau (VHL) tumor suppressor gene is inactivated in both sporadic and inherited clear cell renal carcinoma associated with VHL disease. We have identified two distinct native products of the human VHL gene, with apparent molecular masses of 24 and 18 kDa. The 18-kDa VHL protein was more abundant in nearly all cell lines examined. Reintroduction of the 18-kDa VHL gene product into renal carcinoma cells lacking wild-type VHL protein led to down-regulation of vascular endothelial growth factor (VEGF) mRNA and glucose transporter GLUT1 protein and suppressed tumor formation in nude mice. The 18-kDa VHL protein also demonstrated binding to elongins B and C. In an in vitro assay, the second in-frame AUG codon present in VHL mRNA was shown to be necessary and sufficient for production of the 18-kDa VHL protein, consistent with an internal translation mechanism. These data provide evidence for a second major VHL gene product, which contains the functional domains of the VHL gene. Moreover, these results indicate that internal translation initiation is an important mechanism for production of the major VHL protein.
von Hippel–Lindau (VHL) disease is caused by inactivation of the VHL gene, located on chromosome 3p25–26 (ref. 1, reviewed in ref. 2). Individuals who carry a germ-line mutation in the VHL gene are predisposed to various types of tumors, including renal cell carcinomas, retinal and central nervous system hemangioblastomas, and pheochromocytoma (3, 4). Somatic mutation and/or hypermethylation of the VHL gene have also been implicated in sporadic clear cell renal carcinomas (5–7), facilitating classification of the VHL gene as a tumor suppressor (8).
To understand the mechanisms through which VHL mediates tumor suppression, biochemical analyses of VHL gene products are necessary. However, the translation start site was not evident from the initial cloning of the human VHL gene (1). The first AUG in the human VHL open reading frame (ORF) was designated as the translation start site (amino acid 1) on the basis of homology between the mouse, rat, and human VHL sequences (9, 10), and a product initiated at this start site has been detected in cell lines (11, 12). This site, however, contains a suboptimal Kozak consensus sequence (13). A second AUG is present in the VHL ORF at codon 54. This region contains a more conserved Kozak consensus sequence by virtue of the sequence, AUGG, and thus may serve as a second, internal, translation initiation site.
The importance of a second translation start site is underscored by the lack of mutations found between the first and second methionine codons of the VHL gene in both sporadic and VHL-associated renal carcinomas (4, 5, 14, 15). This observation suggests that mutation in this region might not lead to VHL inactivation if translation could be initiated at the second methionine codon, producing a functional VHL protein. Furthermore, both the rat and mouse contain only 19 of the 53 amino acids present in this region of the human VHL ORF, retaining only one of the eight acidic Gly-Xaa-Glu-Glu-Xaa motifs found in the human sequence (9, 10). Thus, this 53 amino acid region may make little or no contribution to the biological activity of VHL protein.
To address these possibilities, we sought to characterize the native VHL gene products. Here, we report the identification of a second native VHL gene product and demonstrate that this protein is initiated from the second translation start site and contains the biological activity of the VHL gene.
MATERIALS AND METHODS
Cell Culture.
Cell lines 786-0, 293, HeLa, MCF7, and HT-29 were obtained from the American Type Culture Collection (now in Manassas, VA). The CCF-RC2 (16) and U20S cell lines were kindly provided by Jonathan Fleishmann (Albert Einstein College of Medicine) and Liang Zhu (Albert Einstein College of Medicine), respectively. All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS). The 786-0 renal carcinoma cells were transfected in 35-mm culture dishes with 2 μg of DNA and 8 μl of Lipofectamine (Life Technologies, Gaithersburg, MD) in serum-free medium (1 ml). Stably transfected colonies were selected, expanded, and maintained in media containing G418 (600 μg/ml; Life Technologies). For metabolic labeling, cells grown to 95% confluence in a 100-mm culture dish were incubated in methionine-free DMEM with 5% FCS for 1 hr and were subsequently grown for 4 hr in medium containing 500 μCi/ml [35S]methionine (EXPRE35S35S, New England Nuclear; 1 μCi = 37 kBq).
Plasmids.
VHL cDNA vectors, g7 (1) and pSci (containing VHL bases 1–1816, as reported in ref. 1, inserted into HindIII and EcoRI sites in pBluescript II SK (Stratagene), were generously provided by Igor Kuzmin (National Cancer Institute, Frederick, MD) and Michael Lerman (National Cancer Institute, Frederick, MD). Vectors directing expression from either the first or second methionine codon in the VHL coding region were created by PCR amplification of the g7 cDNA by using 5′ primers flanking either start codon and a 3′ primer flanking the stop codon. The amplified DNA fragment was directly ligated into pCR3 (Invitrogen). For translation initiation studies, a construct was created by PCR amplification from the VHL cDNA vector pSci, using the primers 5′-GGAGCGCGCACGAAGCTTCGCCCCGCGTCC-3′ and 5′CAGTGTAAGTTTCTCGAGAAATCTTCAATCTCCCATCCG-3′ (underlined sequences indicate HindIII and XhoI restriction sites, respectively). The PCR product was then digested with HindIII and XhoI and ligated into the cognate sites in pCR3. Mutations in the VHL coding region were introduced by sequential PCR steps using overlapping primers containing the desired point mutations, as described (17). Internal deletion of VHL (amino acids 114–178) was created by PCR amplifications of the two nondeleted fragments, using primers containing SalI restriction sites at the joining ends. The PCR products were digested with SalI, gel purified, and ligated. The ligation mixture was used as template for a second PCR reaction using a 5′ primer flanking the first methionine codon in the VHL coding region and a 3′ primer flanking the stop codon. The resulting product was ligated into pCR3, as described. All plasmids were confirmed by restriction enzyme digestion and DNA sequence analysis.
Antibodies.
Rabbit polyclonal antisera to epitopes encoded within VHL exons 1, 2, and 3 were raised against peptides consisting of amino acids 54–68, 118–129, and 167–177 of the VHL coding region, respectively. For monoclonal antibodies (mAbs), VHL (amino acids 54–213) was expressed as a glutathione S-transferase (GST) fusion protein in BL21(DE3)pLysS cells (Novagen). GST-VHL (amino acids 54–213) was purified on glutathione-agarose, cleaved with thrombin, boiled in 2× SDS gel loading buffer (18), and separated by electrophoresis on an SDS/15% polyacrylamide gel. Proteins were visualized in the gel by incubation in 0.1 M KCl for 5 min. Gel slices containing VHL (amino acids 54–213) were pulverized as described (17), and 50 μg (as determined by direct comparison with concentration standards on a Coomassie blue-stained SDS/PAGE gel) was injected into BALB/c mice. Mice were boosted with 100 μg of soluble GST-VHL (amino acids 1–213) prepared in a similar fashion, except the protein was eluted from the glutathione-agarose beads with glutathione elution buffer (10 mM reduced glutathione/50 mM Tris, pH 8) and protein concentration was determined by the Bradford assay (Bio-Rad). Splenic fusion of lymphocytes from immunized mice with AG8.643 myeloma cells was performed as described (19). Hybridomas were screened by ELISA using 96-well plates coated with GST-VHL (amino acids 1–213) (500 ng per well). Cells from positive wells were cloned in soft agar as described (20). The resulting mAbs, 12.21 and 11E12, recognize epitopes in the carboxyl portion of the VHL protein (data not shown).
Immunoprecipitations.
Cells grown to confluence in 100-mm culture dishes were lysed in 0.5 ml of lysis buffer [50 mM Hepes, pH 7.6/250 mM NaCl/0.1% Nonidet P-40/5 mM EDTA/1 mM phenylmethylsulfonyl fluoride (PMSF) containing 1 μg/ml each of aprotinin, bestatin, and leupeptin] for 30 min at 4°C. Lysates were clarified by centrifugation in a microcentrifuge for 15 min at 4°C. All subsequent incubations and washing steps were done at 4°C. For 35S-labeled lysates, a preclearing step consisting of a 1-hr incubation with normal rabbit serum followed by a 30-min incubation with Zysorbin (Zymed) was added. A 200-μl sample of the clarified lysates was incubated for 1.5 hr with mAb supernatant (100 μl) or ascites fluid (2 μl in 100 μl of lysis buffer) in the presence of 0.67% sodium deoxycholate. Immune complexes were collected on protein A Sepharose (Pharmacia), washed three times with lysis buffer containing 1% Nonidet P-40 (and 500 mM NaCl for 35S-labeled proteins), washed once with lysis buffer, boiled in SDS loading buffer, and separated by SDS/PAGE. Proteins were visualized by Western blotting or by fluorography (for 35S-labeled proteins).
In Vitro Transcription/Translation.
In vitro transcription/translation was performed with TnT coupled reticulocyte lysate system (Promega) according to the manufacturer’s recommendations. For analysis, 15 μl of each reaction was diluted to 200 μl with lysis buffer and immunoprecipitation was performed as previously described.
Peptide Mapping.
Peptide mapping was performed as described (21).
Tumor Suppression.
786-0 renal carcinoma cells were stably transfected with VHLp24(MPR), VHLp18(MEA), or pCR3 (vector only), and expression of the expected VHL products was verified by Western blot analysis. Cells (107) of each cell line were trypsinized, washed, resuspended in PBS, and injected into one flank of a nude mouse. Control cells (untransfected 786-0 and pCR3-transfected 786-0 cells) were injected into the left flank and VHL-expressing cells were injected into the right flank. After 9 weeks, mice were sacrificed and tumors were measured, excised, and weighed.
Binding Assay.
GST and GST-VHL fusion proteins were collected on glutathione-agarose. Beads containing 5 μg of protein, as determined by Coomassie blue staining, were incubated with 20 μl of 35S-labeled elongin B and C products (in vitro transcribed/translated simultaneously from plasmids provided by Arnim Pause, National Institutes of Health, Bethesda, MD) for 1.5 hr at 4°C. Beads were washed three times with lysis buffer containing 500 mM NaCl, washed once with lysis buffer, boiled in 2× SDS gel loading buffer, and separated by electrophoresis on an SDS/8–16% polyacrylamide gradient gel. Proteins were visualized by fluorography.
RESULTS
A protein produced by translation initiation from the first AUG codon in the VHL ORF has previously been identified in human cell lines by immunoprecipitation (11, 12). On the basis of hydropathy plot analysis, a protein initiated from the second translation start site of the VHL ORF was predicted to be more hydrophobic than one initiated from the first AUG codon (data not shown). Thus, we reasoned that the inclusion of strong dissociating agents in the immunoprecipitation step would be necessary to detect a polypeptide translated from the second in-frame AUG. To test this notion, several detergents were assayed for their ability to aid in immunoprecipitation of VHL gene products overexpressed in stably transfected 786-0 renal carcinoma cells (which lack wild-type VHL) from constructs directing translation initiation at either the first or second AUG codon. For clarity, we refer to these two VHL gene products as VHLp24(MPR) and VHLp18(MEA), respectively, by virtue of their apparent molecular mass and first three amino acids (MPR = Met-Pro-Arg; MEA = Met-Glu-Ala). When a monoclonal antibody (mAb) to VHL, mAb 12.21, was used and immune complexes were visualized by Western blotting, sodium deoxycholate (0.67%) proved to be the most effective in facilitating immunoprecipitation of both VHLp24(MPR) and VHLp18(MEA) products (Fig. 1, lanes 1 and 2). VHLp24(MPR) appears as at least two bands, possibly representing post-translational modification.
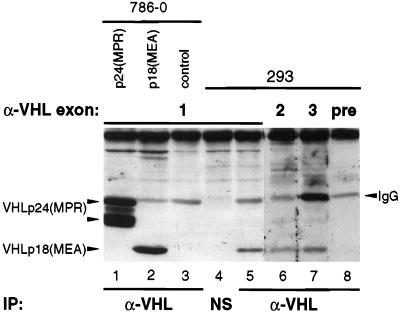
Figure 1.
Identification of native VHL gene products. Total cellular extracts from 786-0 cells stably expressing VHLp24(MPR) (lane 1), VHLp18(MEA) (lane 2), or control 786-0 cells (lane 3) or from 293 cells (lanes 4–8) were immunoprecipitated (IP) with either VHL mAb 12.21 (lanes 1–3, 5–8), or a nonspecific (unrelated) mAb, 12CA5 (NS, lane 4). Immunoprecipitates were separated by electrophoresis on an SDS/12.5% polyacrylamide gel and transferred to a poly(vinylidene difluoride) (PVDF) membrane. The membrane was cut and Western blotting was performed on the pieces by using rabbit polyclonal antisera to peptides in VHL exon 1 (lanes 1–5), exon 2 (lane 6), exon 3 (lane 7), or preimmune rabbit sera (lane 8). VHLp24(MPR) and VHLp18(MEA) are indicated to the left of the blot. Cross-reacting IgG (light chain) is indicated to the right of the blot. NS mAb (lane 4) is subtype IgG2b and did not cross-react with the anti-rabbit IgG secondary antibody.
When native VHL products from the human embryonic kidney cell line, 293, were analyzed by immunoprecipitation/Western blotting, a protein that comigrates with VHLp18(MEA) was evident (Fig. 1, lane 5); this protein was considerably more abundant than the slower-migrating VHLp24(MPR). The protein comigrating with VHLp18(MEA) was not detected when extracts from 293 cells were immunoprecipitated with an unrelated mAb (Fig. 1, lane 4). Similar results were obtained by immunoprecipitation with additional polyclonal and monoclonal VHL antibodies (data not shown).
To rule out the possibility that the faster-migrating native protein was caused by alternative splicing of the VHL gene, cellular extracts from 293 cells were immunoprecipitated with mAb 12.21 and the resulting complexes were immunoblotted using rabbit polyclonal antisera to each of the three exons in the VHL ORF (Fig. 1, lanes 5–7). The endogenous protein comigrating with VHLp18(MEA) was detected by antibodies to each of the three exons, but not with rabbit preimmune serum (Fig. 1, lane 8).
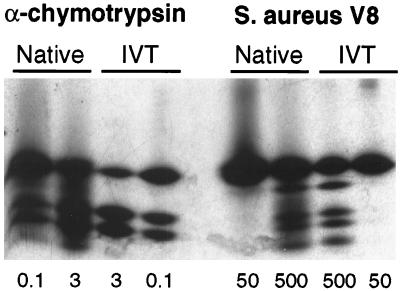
To confirm that the VHL product migrating at 18 kDa was, in fact, VHLp18(MEA), peptide mapping of this endogenous protein was performed. 293 cells were metabolically labeled with [35S]methionine and immunoprecipitated with mAb 11E12. Immune complexes were visualized by fluorography and the native VHL protein, as identified by comigration with in vitro translated VHLp18(MEA), was excised from the gel. Partial proteolysis of the isolated protein with either α-chymotrypsin or Staphylococcus aureus V8 protease produced peptide maps that were identical to those of in vitro translated and digested VHLp18(MEA) (Fig. 2). These results establish that this endogenous protein is, indeed, VHLp18(MEA).
Figure 2.
Peptide mapping of VHLp18(MEA). Native VHL products were immunoprecipitated from metabolically labeled 293 cells, separated on an SDS/13.5% polyacrylamide gel, and visualized by fluorography. Bands comigrating with in vitro translated VHLp18(MEA) were excised from the gel and rehydrated. Partial proteolysis was performed (21) on native and in vitro translated products (IVT) by using amounts of α-chymotrypsin (μg) or S. aureus V8 protease (ng) indicated below the figure. Digestion products were separated on an SDS/15% polyacrylamide gel and visualized by fluorography.
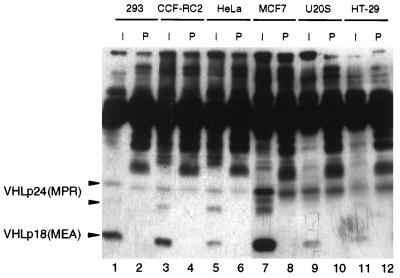
We then sought to determine whether VHLp18(MEA) was present in other human cell lines. In addition to 293 cells, CCF-RC2 (renal carcinoma), HeLa (cervical carcinoma), MCF7 (breast adenocarcinoma), U20S (osteosarcoma), and HT-29 (colon adenocarcinoma) cell lines were assayed by immunoprecipitation/Western blotting (Fig. 3). VHLp18(MEA) was the predominant VHL immunoreactive protein detected in 293, CCF-RC2, MCF7, U20S, and HT-29 cell lines (Fig. 3, lanes 1, 3, 7, 9, 11) and was equally abundant with VHLp24(MPR) in HeLa cells (Fig. 3, lane 5).
Figure 3.
Detection of VHL gene products in cell lines. Immunoprecipitations were performed on indicated cell lines by using either VHL mAb 11E12 (I) or mouse preimmune serum (P), as indicated above lanes. Immune complexes were separated on an SDS/13.5% polyacrylamide gel, transferred to poly(vinylidene difluoride) membrane, and immunoblotted with mAb 12.21. Positions of VHLp24(MPR) and VHLp18(MEA) are indicated to the left of the blot.
Next, we sought to determine whether VHLp18(MEA) has the ability to affect cell growth and/or suppress tumor formation. 786-0 renal carcinoma cells (which lack wild-type VHL) were stably transfected with pCR3 (vector alone), VHLp24(MPR), or VHLp18(MEA) plasmids. Multiple independent clones were obtained for each construct and VHL protein expression was documented by Western blotting (Fig. 1, lanes 1 and 2 and data not shown). The expression of VHLp18(MEA) in 786-0 cells had no effect on their growth properties in plastic tissue culture dishes. Population doubling time, cell cycle progression, and cell morphology were identical for untransfected cells and cells expressing VHLp18(MEA) (data not shown).
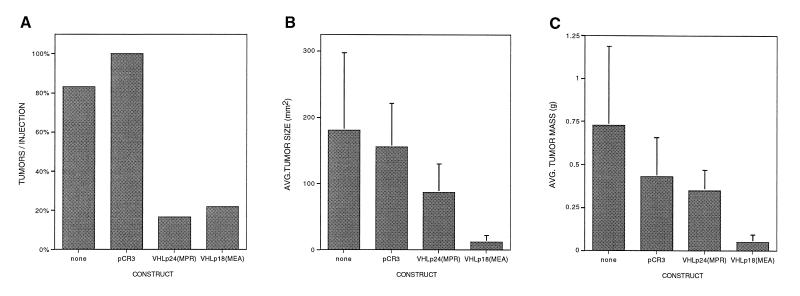
The stably transfected clones were then assayed for tumor formation in nude mice as described in Materials and Methods. In total, 66 mouse flanks were injected: 18 with VHLp24(MPR) (three independent clones repeated six times), 18 with VHLp18(MEA) (three independent clones repeated six times), 18 with pCR3 (two independent clones repeated twelve and six times), and 12 with untransfected 786-0 cells. Both VHLp18(MEA) and VHLp24(MPR) effectively suppressed tumor formation as compared with control cells (pCR3-containing clones or untransfected 786-0 cells) (P < 0.001, χ2 test) (Fig. 4A). The number of tumors formed per injection was equivalent for VHLp18(MEA) and VHLp24(MPR) (4/18 vs. 3/18). The tumors formed in the presence of VHLp18(MEA) were significantly smaller in size and mass than those of VHLp24(MPR) (P < 0.05 and P < 0.01, respectively, Student’s t test) (Fig. 4 B and C). Thus, the VHLp18(MEA) protein contains the domains necessary for tumor suppressor function.
Figure 4.
Tumor suppression by VHL gene products. Colonies of 786-0 renal carcinoma cells, stably transfected with the indicated plasmids, were assayed for tumor formation in nude mice. Cells (107) of each cell line were trypsinized, washed, resuspended in PBS, and injected into one flank of a nude mouse. Control cells (untransfected 786-0 and pCR3) were injected into the left flank and VHL-expressing cells were injected into the right flank. After 9 weeks, mice were sacrificed and tumors were measured, excised, and weighed. (A) Tumors per injection. Bars represent the number of tumors as a percentage of total injections for each construct [total injections = 18 for pCR3, VHLp24(MPR), and VHLp18(MEA) and 12 for untransfected 786-0 cells]. Three independent clones of VHLp24(MPR) and VHLp18(MEA) and two independent clones of pCR3 were assayed. (B) Average size (mm2) of detected tumors for each construct. (C) Average mass (g) of detected tumors for each construct.
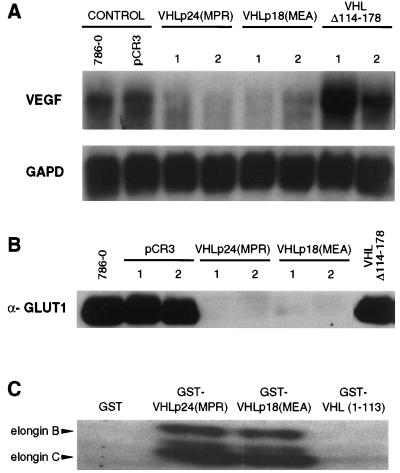
We then sought to determine whether the tumor suppressor function of VHLp18(MEA) was associated with negative regulation of vascular endothelial growth factor (VEGF) and the glucose transporter, GLUT1, as well as elongin B and C binding, as previously shown for VHLp24(MPR) (12, 22–26). Similar to down-regulation seen in VHLp24(MPR)-expressing cells, down-regulation of VEGF mRNA (as analyzed by Northern blotting) was seen in cells that express VHLp18(MEA) (Fig. 5A). GLUT1 down-regulation by VHLp18(MEA), assayed at the protein level, was also identical to that observed for VHLp24(MPR) (Fig. 5B). Down-regulation of VEGF and GLUT1 did not occur in cells expressing a mutant VHL (VHL Δ114–178, Fig. 5 A and B), however. In addition, a GST-VHLp18(MEA) fusion protein bound to in vitro translated elongins B and C to an extent equal to that of GST-VHLp24(MPR) (Fig. 5C), whereas neither GST alone nor a mutant GST-VHL (amino acids 1–113) demonstrated binding. Likewise, VHLp18(MEA) binding to cul-2 has been demonstrated previously (27). Thus, VHLp18(MEA) possesses important VHL biological activities.
Figure 5.
Down-regulation of VEGF and GLUT1 by VHL gene products. 786-0 cells, either untransfected or stably transfected with pCR3, VHLp24(MPR), VHLp18(MEA), or VHL Δ114–178 were assayed for VEGF RNA (A) and GLUT1 protein (B). (A) Northern blot for VEGF. Total RNA was isolated from cell lines (indicated above blot) grown to low confluence (50–70%), and 40 μg of total RNA was loaded per well. Two independent clones of VHLp24(MPR), VHLp18(MEA), and VHL Δ114–178 were assayed. After probing with VEGF, the blot was stripped and rehybridized with a glyceraldehyde-3-phosphate dehydrogenase (GAPD) probe. Probes are indicated to the left of the blot. (B) Anti-GLUT1 Western blot. Each lane was loaded with 25 μg of protein extract from cell lines (indicated above blot). Western blotting was performed with GLUT1 antisera (Alpha Diagnostic International, San Antonio, TX). Two independent clones of pCR3, VHLp24(MPR), and VHLp18(MEA) were assayed. (C) Elongin binding assay. Beads containing 5 μg of GST or GST-VHL fusion proteins (indicated above lanes) were incubated with 20 μl of in vitro translated elongins B and C, washed, separated on an SDS/8–16% polyacrylamide gel, and visualized by fluorography. Positions of the elongin B and C products are indicated to the left.
Next, we examined the cellular mechanisms responsible for the production of two VHL gene products in 293 cells. The transcription start sites of the VHL gene have been extensively mapped in 293 cells (28), and all transcripts detected contain the first AUG codon. These results suggest that the production of VHLp18(MEA) in vivo is caused either by translation initiation from the second in-frame AUG in the VHL coding region or by processing of the VHLp24(MPR) product. Because VHLp18(MEA) was not detected in 786-0 cells that overexpress VHLp24(MPR) (see Fig. 1, lanes 1 and 2), VHLp18(MEA) production by partial degradation is unlikely.
On the basis of the high G+C content (81%) and extensive secondary structure in the 5′ region of the native mRNA predicted by computer analyses (data not shown), we reasoned that use of the first translation start codon might be hindered, leading to translation initiation at the second AUG codon (29). To test this possibility, we created an expression vector that reproduces the 5′ end of a mapped native VHL transcript (28) (described in Materials and Methods) when T7 RNA polymerase is used to direct transcription from the T7 promoter region. This construct, VHLNT (native transcript), produces an mRNA that is initiated three bases upstream (at −8, according to numbering in ref. 28) of the first mapped nucleotide of native VHL mRNA (at −5). Furthermore, this mRNA retains the native VHL 5′ untranslated sequence, differing by only one base (at +4, because of an engineered restriction enzyme site) between the major transcription start site (+1) and the first translation start site (+68).
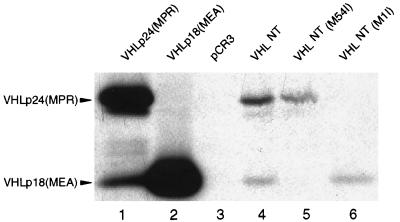
The products of in vitro transcription/translation reactions using VHLNT as template were compared with those of VHLp24(MPR) and VHLp18(MEA) constructs containing heterologous 5′ untranslated sequences (see Fig. 1, lanes 1 and 2). Both VHLp24(MPR) and VHLp18(MEA) constructs produce copious amounts of their expected products (Fig. 6, lanes 1 and 2). Surprisingly, the VHLp24(MPR) construct also yielded a lesser, yet substantial, amount of VHLp18(MEA) product. The VHLNT construct was also able to produce both VHLp24(MPR) and VHLp18(MEA) proteins (Fig. 6, lane 4), although in lower abundance, presumably because of the inhibitory effect of its mRNA secondary structure. When the second AUG codon of VHLNT was changed to AUU (causing a substitution of isoleucine for methionine), the resulting construct, VHLNT(M54I), was unable to produce VHLp18(MEA) (Fig. 6, lane 5). This outcome demonstrates that the VHLp18(MEA) product is not a breakdown product of the VHLp24(MPR) protein but, in fact, is a result of translation initiation at this second internal start codon. Mutation of the first initiation codon in the construct VHLNT(M1I) ablated expression of VHLp24(MPR) but failed to produce higher levels of VHLp18(MEA) product (Fig. 6, lane 6), as would be expected if VHLp18(MEA) were produced by leaky ribosomal scanning (29). Addition of the native VHL 3′ untranslated region had no effect on either VHLp24(MPR) or VHLp18(MEA) production by VHLNT (data not shown), excluding any contribution of the heterologous 3′ untranslated region present in VHLNT in this system. Similar results were also obtained by transfection of these constructs into 786-0 cells that had been infected with a recombinant vaccinia virus that expresses T7 RNA polymerase (30) (data not shown).
Figure 6.
In vitro translation of VHL gene products. In vitro transcription/translation was performed in the presence of [35S]methionine. Heterologous leader sequence plasmids VHLp24(MPR), VHLp18(MEA), and pCR3 and native 5′ untranslated sequence constructs VHLNT, VHLNT(M54I), and VHLNT(M1I) were used as templates (as indicated above the lanes). Fifteen microliters of the resulting products was immunoprecipitated with mAb 11E12, separated on an SDS/13.5% polyacrylamide gel, and visualized by fluorography. Positions of the VHLp24(MPR) and VHLp18(MEA) products are indicated to the left.
DISCUSSION
These results demonstrate that in addition to the previously detected product of the VHL gene, VHLp24(MPR), there also exists a second native product, VHLp18(MEA), which is created by translation initiation from an internal start site. In our studies, the overall level of native VHLp18(MEA) product detected in cell lines was generally higher than that of VHLp24(MPR), differing from previous studies (11, 12). Because VHLp18(MEA) lacks the acidic N-terminal region that is present in VHLp24(MPR), it is more hydrophobic and requires the use of strong dissociating agents such as deoxycholate to free it for antibody binding. Therefore, lack of adequate detergents (e.g., deoxycholate) in the immunoprecipitation step leads to an underestimation of the amount of VHLp18(MEA) in cells. Thus, the present results more accurately reflect the relative abundance of native VHL products in cell lines.
VHLp18(MEA) suppressed tumor formation in nude mice. However, the expression of VHLp18(MEA) in 786-0 cells had no phenotypic effect on the growth of these cells in culture on plastic, similar to results of studies of the tumor suppressor properties of VHLp24(MPR) (11). Thus, the tumor suppressor ability of VHLp18(MEA) was not a nonspecific toxic effect of overexpression on the cells. Also supporting this idea, a VHL truncation mutant consisting of amino acids 1–115 was unable to block the formation of tumors in nude mice (11).
Because both VHLp24(MPR) and VHLp18(MEA) gene products have similar biological effects such as tumor suppressor ability, down-regulation of VEGF and GLUT1, and elongin B and C binding, this might point to a common mechanism of action. However, it is possible that VHLp24(MPR) and VHLp18(MEA) might also have independent activities. This idea is plausible, as these two products have been shown to have different subcellular localizations (ref. 31 and A.S., unpublished data). Thus, the existence of two VHL gene products is anticipated to have biological significance.
In this report, we demonstrate that VHLp18(MEA) can be translated from an internal start site in the VHL transcript. The in vitro assay yields lower levels of VHLp18(MEA) product than VHLp24(MPR), differing from the levels detected in cell lines. This difference may be due to one or more cellular translation initiation regulatory factors that are absent from the in vitro lysate. Alternatively, VHLp18(MEA) and VHLp24(MPR) proteins may be degraded at different rates, influencing the in vivo ratio of these products. Also, while this study focuses on the longest native mRNA, the use of other mapped transcription start sites in the VHL gene (all containing the first AUG codon) (28) may favor production of VHLp18(MEA). Our data definitively show that VHLp18(MEA) is not produced by partial degradation of the VHLp24(MPR) protein, as evidenced by the absence of a VHLp18(MEA) product both in 786-0 cells stably expressing VHLp24(MPR) and in in vitro and transient transfection assays performed with a construct containing a mutation of the second methionine codon.
Internal initiation of translation has been demonstrated for the human Ig heavy-chain binding protein, BiP (32), Drosophila Antennapedia (33), and human fibroblast growth factor 2 (FGF-2) (34) mRNAs. It is interesting that FGF-2, an angiogenic factor involved in tumor neovascularization (35), and VHL, which (by means of VEGF down-regulation) is anti-angiogenic, both utilize internal translation initiation. Thus, an understanding of the biological consequences of these alternative translation mechanisms may provide further insight into the function of the VHL gene and its role in suppressing tumorigenesis.
Acknowledgments
We thank Umadas Maitra, Charles E. Rogler, and David A. Shafritz for critical review of this manuscript. R.B. thanks the instructors of the 1995 Cold Spring Harbor course in Protein Purification and Characterization for their time and assistance. DNA oligonucleotides were synthesized in the oligonucleotide facility of the Cancer Center of the Albert Einstein College of Medicine (partially supported by Grant CA 13330). mAbs were produced at the Hybridoma Facility of the Cancer Center of the Albert Einstein College of Medicine (partially supported by Grant CA 13330). A.S. is supported by a National Institutes of Health Training grant (CA 09060). E.D. is supported by a National Institutes of Health Training Grant (DK 07218).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: VHL, von Hippel–Lindau; GST, glutathione S-transferase; VEGF, vascular endothelial growth factor; MPR, Met-Pro-Arg; MEA, Met-Glu-Ala; NT, native transcript.
References
- 1.Latif F, Tory K, Gnarra J, Yao M, Duh F M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 2.Linehan W M, Lerman M I, Zbar B. J Am Med Assoc. 1995;273:564–570. [PubMed] [Google Scholar]
- 3.Maher E R, Iselius L, Yates J R, Littler M, Benjamin C, Harris R, Sampson J, Williams A, Ferguson S M A, Morton N. J Med Genet. 1991;28:443–447. doi: 10.1136/jmg.28.7.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gnarra J R, Tory K, Weng Y, Schmidt L, Wei M H, Li H, Latif F, Liu S, Chen F, Duh F M, et al. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 5.Whaley J M, Naglich J, Gelbert L, Hsia Y E, Lamiell J M, Green J S, Collins D, Neumann H P, Laidlaw J, Li F P, et al. Am J Hum Genet. 1994;55:1092–1102. [PMC free article] [PubMed] [Google Scholar]
- 6.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D R, Gnarra J R, Linehan W M, et al. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crossey P A, Foster K, Richards F M, Phipps M E, Latif F, Tory K, Jones M H, Bentley E, Kumar R, Lerman M I, et al. Hum Genet. 1994;93:53–58. doi: 10.1007/BF00218913. [DOI] [PubMed] [Google Scholar]
- 8.Knudson A G. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Naglich J G, Laidlaw J, Whaley J M, Seizinger B R, Kley N. Cancer Res. 1995;55:743–747. [PubMed] [Google Scholar]
- 10.Duan D R, Humphrey J S, Chen D Y, Weng Y, Sukegawa J, Lee S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1995;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos O, Kibel A, Gray S, Kaelin W G. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 12.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Kishida T, Yao M, Hustad T, Glavac D, Dean M, Gnarra J R, Orcutt M L, Duh F M, Glenn G, et al. Hum Mutat. 1995;5:66–75. doi: 10.1002/humu.1380050109. [DOI] [PubMed] [Google Scholar]
- 15.Prowse A H, Webster A R, Richards F M, Richard S, Olschwang S, Resche F, Affara N A, Maher E R. Am J Hum Genet. 1997;60:765–771. [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimura T, Tubbs R R, Connelly R, Caulfield M J, Trindade C S, McMahon J T, Galetti T P, Edinger M, Sandberg A A, Cin D P, Sait S J, Pontes J E. Cancer Res. 1989;49:7064–7071. [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, Janssen K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Fazekas de St. Groth S, Scheidegger D. J Immunol Methods. 1980;35:1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- 20.French D, Kelly T, Buhl S, Scharff M D. Methods Enzymol. 1987;151:50–66. doi: 10.1016/s0076-6879(87)51008-4. [DOI] [PubMed] [Google Scholar]
- 21.Cleveland D, Fischer S, Kirschner M, Laemmli U. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 22.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Goldberg M A. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassiliou E, Oldfield E H, Klausner R D, Linehan W M. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy A P, Levy N S, Goldberg M A. J Biol Chem. 1996;271:25492–25497. doi: 10.1074/jbc.271.41.25492. [DOI] [PubMed] [Google Scholar]
- 25.Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 26.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 27.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaelin W G. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuzmin I, Duh F-M, Latif F, Geil L, Zbar B, Lerman M I. Oncogene. 1995;10:2185–2194. [PubMed] [Google Scholar]
- 29.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuerst T R, Niles G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Chen D Y T, Humphrey J S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macejak D G, Sarnow P. Nature (London) 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 33.Oh S K, Scott M P, Sarnow P. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 34.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandel J, Bossy-Wetzel E, Radvanyi F, Klagsbrun M, Folkman J, Hanahan D. Cell. 1991;66:1095–1104. doi: 10.1016/0092-8674(91)90033-u. [DOI] [PubMed] [Google Scholar]