Abstract
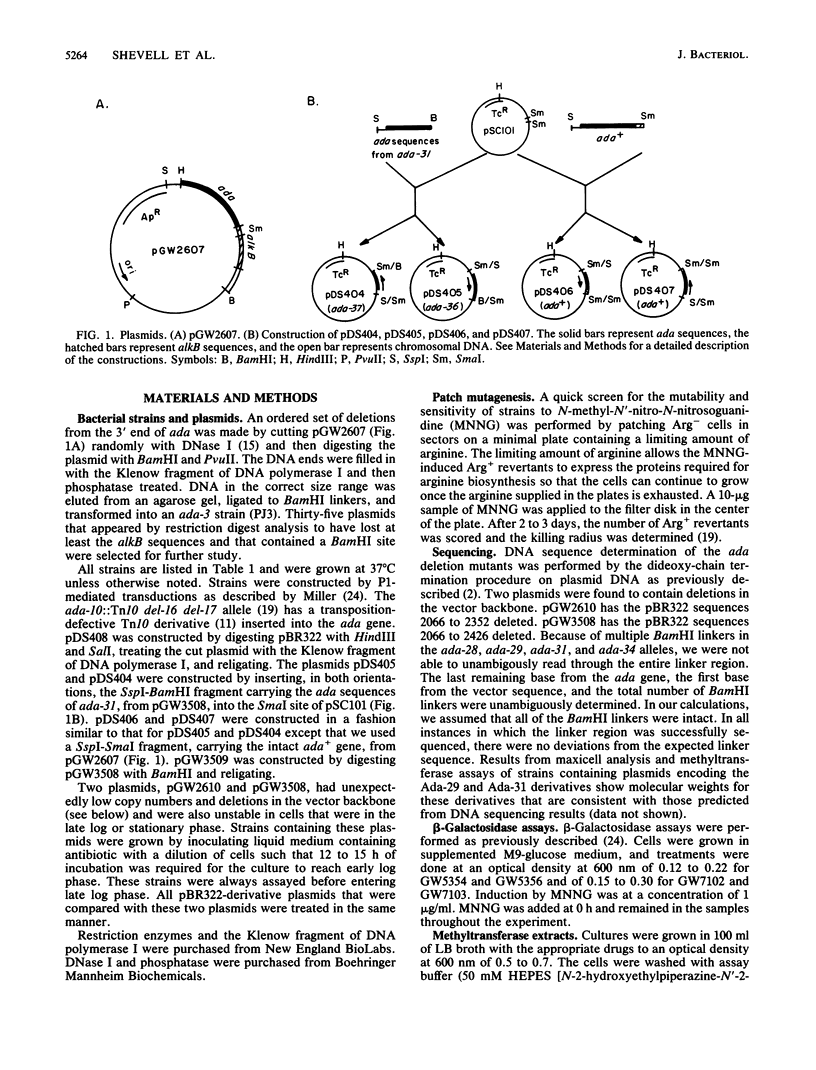
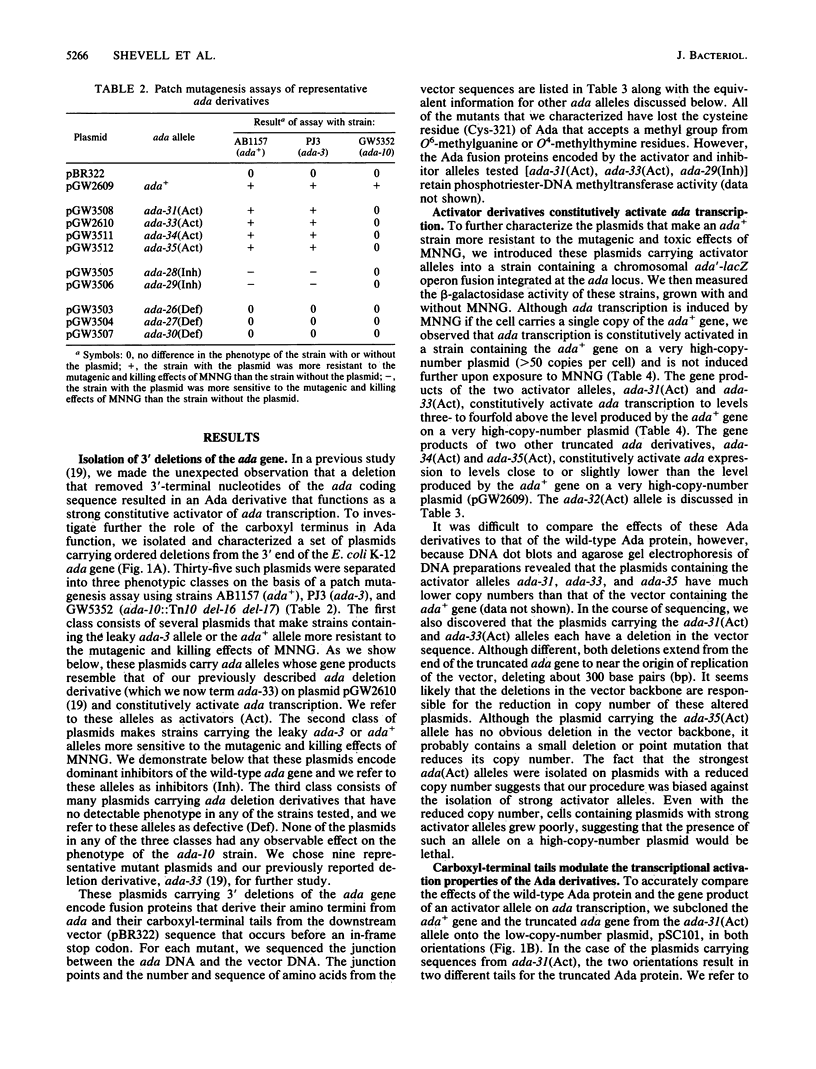
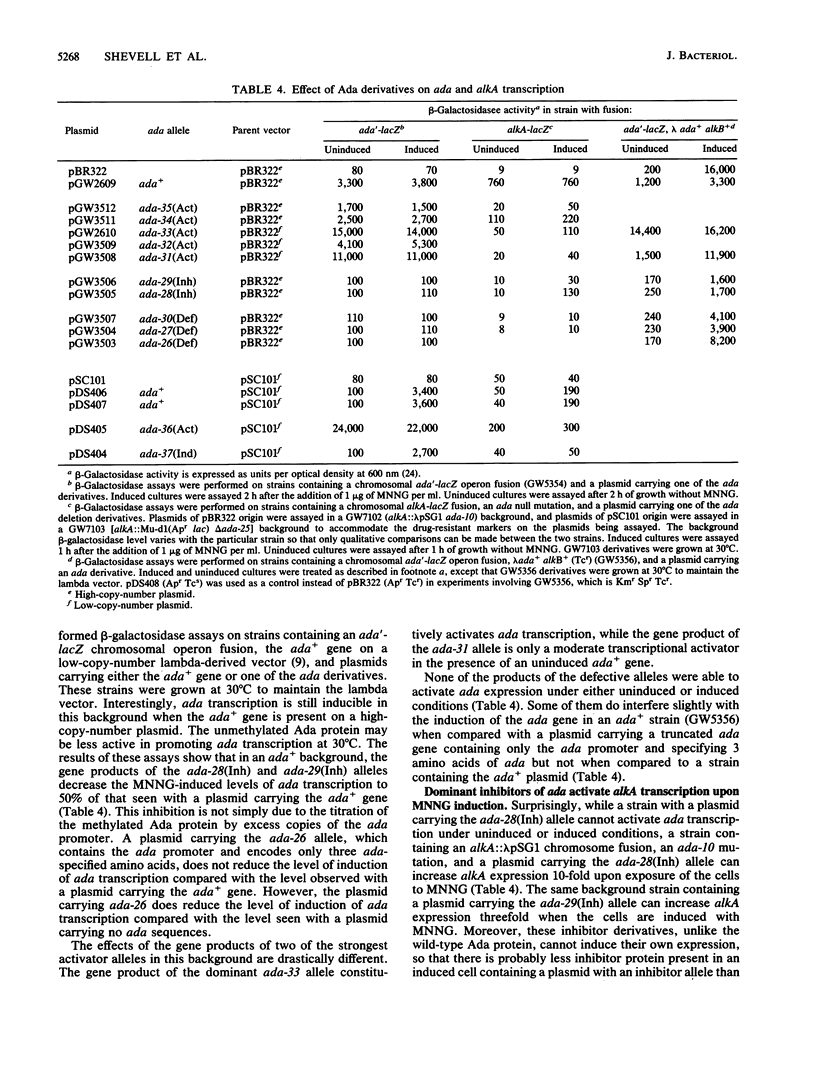
The ada gene of Escherichia coli K-12 encodes the regulatory protein for the adaptive response to alkylating agents. A set of plasmids carrying ordered deletions from the 3' end of the ada gene were isolated and characterized. These ada deletions encode fusion proteins that derive their amino termini from ada and their carboxyl termini from the downstream vector sequence that occurs before an in-frame stop codon. Several of these ada deletions encode Ada derivatives that constitutively activate ada transcription to very high levels. A second class of ada deletions encode Ada derivatives that are dominant inhibitors of the inducible transcription of ada but are inducible activators of alkA transcription. In addition, we found that two Ada derivatives containing the same ada sequences but fused to different vector-derived tails have strikingly different properties. One Ada derivative constitutively activates both ada and alkA expression to very high levels. In contrast, the other Ada derivative is an inducible activator of ada expression, like the wild-type Ada protein, but is not an inducible activator of alkA transcription. Our data suggest that the carboxyl terminus of the Ada protein plays a key role in modulating the ability of the Ada protein to function as a transcriptional activator.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cesareni G., Muesing M. A., Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6313–6317. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Kvaal M., Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197(3):368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- Demple B., Jacobsson A., Olsson M., Robins P., Lindahl T. Repair of alkylated DNA in Escherichia coli. Physical properties of O6-methylguanine-DNA methyltransferase. J Biol Chem. 1982 Nov 25;257(22):13776–13780. [PubMed] [Google Scholar]
- Demple B. Mutant Escherichia coli Ada proteins simultaneously defective in the repair of O6-methylguanine and in gene activation. Nucleic Acids Res. 1986 Jul 25;14(14):5575–5589. doi: 10.1093/nar/14.14.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Phasmid vectors for identification of genes by complementation of Escherichia coli mutants. J Bacteriol. 1985 May;162(2):777–783. doi: 10.1128/jb.162.2.777-783.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Grossman A. D., Erickson J. W., Gross C. A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984 Sep;38(2):383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979 Sep;139(3):783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasen I., Evensen G., Seeberg E. Amplified expression of the tag+ and alkA+ genes in Escherichia coli: identification of gene products and effects on alkylation resistance. J Bacteriol. 1986 Nov;168(2):642–647. doi: 10.1128/jb.168.2.642-647.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H., Sekiguchi M. Molecular cloning and characterization of the alkB gene of Escherichia coli. Mol Gen Genet. 1985;198(2):263–269. doi: 10.1007/BF00383004. [DOI] [PubMed] [Google Scholar]
- Lemotte P. K., Walker G. C. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985 Mar;161(3):888–895. doi: 10.1128/jb.161.3.888-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margison G. P., Cooper D. P., Brennand J. Cloning of the E. coli O6-methylguanine and methylphosphotriester methyltransferase gene using a functional DNA repair assay. Nucleic Acids Res. 1985 Mar 25;13(6):1939–1952. doi: 10.1093/nar/13.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Karran P., Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984 Mar;3(3):545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985 Apr 25;13(8):2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Kondo H., Kawabata S., Iwanaga S., Sekiguchi M. Purification and structure of the intact Ada regulatory protein of Escherichia coli K12, O6-methylguanine-DNA methyltransferase. J Biol Chem. 1985 Jun 25;260(12):7281–7288. [PubMed] [Google Scholar]
- Nakabeppu Y., Kondo H., Sekiguchi M. Cloning and characterization of the alkA gene of Escherichia coli that encodes 3-methyladenine DNA glycosylase II. J Biol Chem. 1984 Nov 25;259(22):13723–13729. [PubMed] [Google Scholar]
- Nakabeppu Y., Mine Y., Sekiguchi M. Regulation of expression of the cloned ada gene in Escherichia coli. Mutat Res. 1985 Sep;146(2):155–167. doi: 10.1016/0167-8817(85)90006-9. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984 Nov 25;259(22):13730–13736. [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Reznikoff W. S. Lactose promoter mutation Pr115 activates an overlapping promoter within the lactose control region. J Mol Biol. 1985 Oct 5;185(3):525–533. doi: 10.1016/0022-2836(85)90069-5. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter P. M., Wilkinson M. C., Fitton J., Carr F. J., Brennand J., Cooper D. P., Margison G. P. Characterisation and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E. coli. Nucleic Acids Res. 1987 Nov 25;15(22):9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck G. W., Coons S., Carroll P., Samson L. A second DNA methyltransferase repair enzyme in Escherichia coli. Proc Natl Acad Sci U S A. 1988 May;85(9):3039–3043. doi: 10.1073/pnas.85.9.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol Gen Genet. 1983;191(3):466–472. doi: 10.1007/BF00425764. [DOI] [PubMed] [Google Scholar]
- Sedgwick B., Robins P., Totty N., Lindahl T. Functional domains and methyl acceptor sites of the Escherichia coli ada protein. J Biol Chem. 1988 Mar 25;263(9):4430–4433. [PubMed] [Google Scholar]
- Shevell D. E., Abou-Zamzam A. M., Demple B., Walker G. C. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J Bacteriol. 1988 Jul;170(7):3294–3296. doi: 10.1128/jb.170.7.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Takano K., Nakabeppu Y., Sekiguchi M. Functional sites of the Ada regulatory protein of Escherichia coli. Analysis by amino acid substitutions. J Mol Biol. 1988 May 20;201(2):261–271. doi: 10.1016/0022-2836(88)90137-4. [DOI] [PubMed] [Google Scholar]
- Teo I. A. Proteolytic processing of the Ada protein that repairs DNA O6-methylguanine residues in E. coli. Mutat Res. 1987 Mar;183(2):123–127. doi: 10.1016/0167-8817(87)90054-x. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Volkert M. R., Nguyen D. C., Beard K. C. Escherichia coli gene induction by alkylation treatment. Genetics. 1986 Jan;112(1):11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Nguyen D. C. Induction of specific Escherichia coli genes by sublethal treatments with alkylating agents. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4110–4114. doi: 10.1073/pnas.81.13.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Weinfeld M., Drake A. F., Saunders J. K., Paterson M. C. Stereospecific removal of methyl phosphotriesters from DNA by an Escherichia coli ada+ extract. Nucleic Acids Res. 1985 Oct 11;13(19):7067–7077. doi: 10.1093/nar/13.19.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]



