Abstract
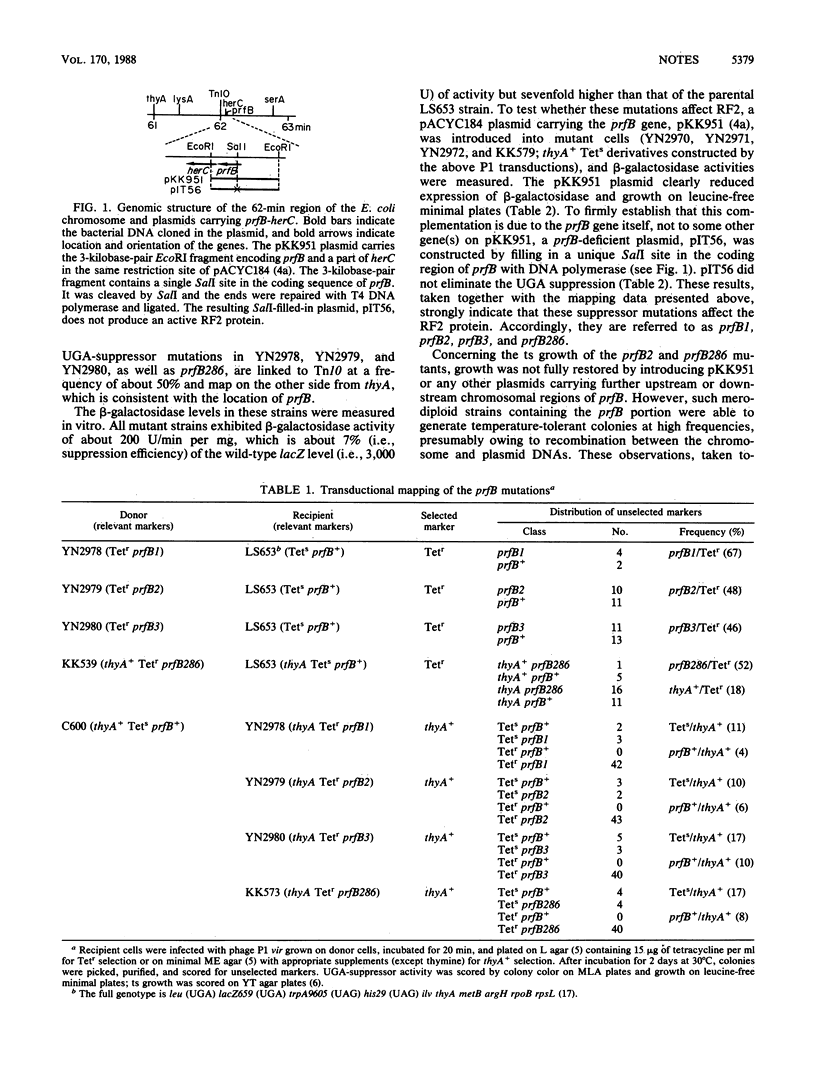
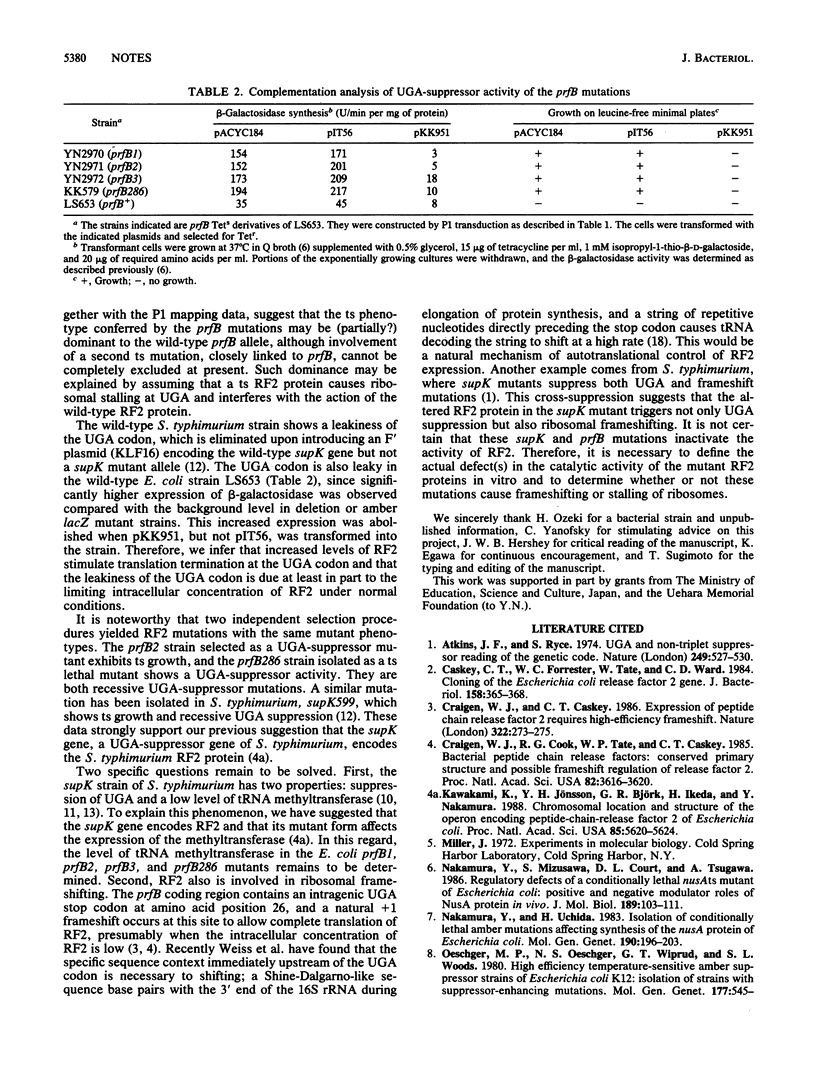
Strains carrying mutations in the prfB gene encoding peptide chain release factor 2 of Escherichia coli were isolated. prfB1, prfB2, and prfB3 were selected as suppressor mutations of a lacZ (UGA) mutation at 37 degrees C, one of which, prfB2, is temperature sensitive in growth. A prfB286 strain was selected as a conditionally lethal mutant which grows at 32 but not at 43 degrees C and was shown to have UGA-suppressor activity. All the mutations are recessive UGA-suppressors. These data indicate that release factor 2 is essential to E. coli growth and that all mutants isolated here trigger suppression of the UGA codon.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Ryce S. UGA and non-triplet suppressor reading of the genetic code. Nature. 1974 Jun 7;249(457):527–530. doi: 10.1038/249527a0. [DOI] [PubMed] [Google Scholar]
- Caskey C. T., Forrester W. C., Tate W., Ward C. D. Cloning of the Escherichia coli release factor 2 gene. J Bacteriol. 1984 Apr;158(1):365–368. doi: 10.1128/jb.158.1.365-368.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Jönsson Y. H., Björk G. R., Ikeda H., Nakamura Y. Chromosomal location and structure of the operon encoding peptide-chain-release factor 2 of Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5620–5624. doi: 10.1073/pnas.85.15.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Mizusawa S., Court D. L., Tsugawa A. Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli. Positive and negative modulator roles of NusA protein in vivo. J Mol Biol. 1986 May 5;189(1):103–111. doi: 10.1016/0022-2836(86)90384-0. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Uchida H. Isolation of conditionally lethal amber mutations affecting synthesis of the nusA protein of Escherichia coli. Mol Gen Genet. 1983;190(2):196–203. doi: 10.1007/BF00330640. [DOI] [PubMed] [Google Scholar]
- Oeschger M. P., Oeschger N. S., Wiprud G. T., Woods S. L. High efficiency temperature-sensitive amber suppressor strains of Escherichia coli K12: isolation of strains with suppressor-enhancing mutations. Mol Gen Genet. 1980;177(4):545–552. doi: 10.1007/BF00272662. [DOI] [PubMed] [Google Scholar]
- Oeschger M. P., Wiprud G. T. High efficiency temperature-sensitive amber suppressor strains of Escherichia coli K12: construction and characterization of recombinant strains with suppressor-enhancing mutations. Mol Gen Genet. 1980;178(2):293–299. doi: 10.1007/BF00270475. [DOI] [PubMed] [Google Scholar]
- Pope W. T., Brown A., Reeves R. H. The identification of the tRNA substrates for the supK tRNA methylase. Nucleic Acids Res. 1978 Mar;5(3):1041–1057. doi: 10.1093/nar/5.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W. T., Reeves R. H. Purification and characterization of a tRNA methylase from Salmonella typhimurium. J Bacteriol. 1978 Oct;136(1):191–200. doi: 10.1128/jb.136.1.191-200.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. H., Roth J. R. A recessive UGA suppressor. J Mol Biol. 1971 Mar 28;56(3):523–533. doi: 10.1016/0022-2836(71)90399-8. [DOI] [PubMed] [Google Scholar]
- Reeves R. H., Roth J. R. Transfer ribonucleic acid methylase deficiency found in UGA supressor strains. J Bacteriol. 1975 Oct;124(1):332–340. doi: 10.1128/jb.124.1.332-340.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden M., Murphy J., Martin R., Isaksson L., Gallant J. Mapping and complementation studies of the gene for release factor 1. J Bacteriol. 1986 Dec;168(3):1066–1069. doi: 10.1128/jb.168.3.1066-1069.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydén S. M., Isaksson L. A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193(1):38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- Scolnick E., Tompkins R., Caskey T., Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968 Oct;61(2):768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll L. Mutational alterations of tryptophan-specific transfer RNA that generate translation suppressors of the UAA, UAG and UGA nonsense codons. J Mol Biol. 1974 Jun 25;86(2):233–243. doi: 10.1016/0022-2836(74)90015-1. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Murphy J. P., Gallant J. A. Genetic screen for cloned release factor genes. J Bacteriol. 1984 Apr;158(1):362–364. doi: 10.1128/jb.158.1.362-364.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]



